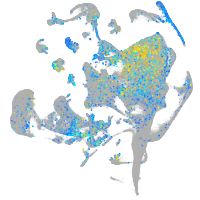
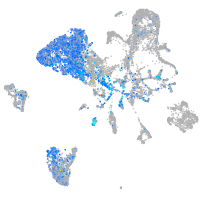
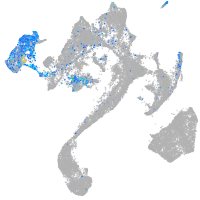
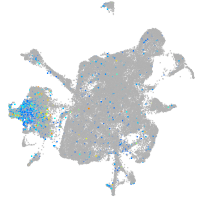
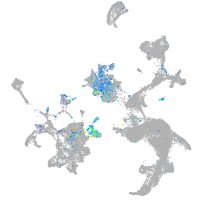
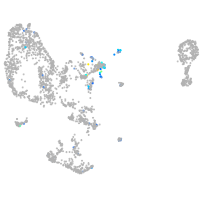
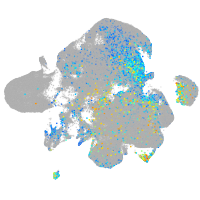
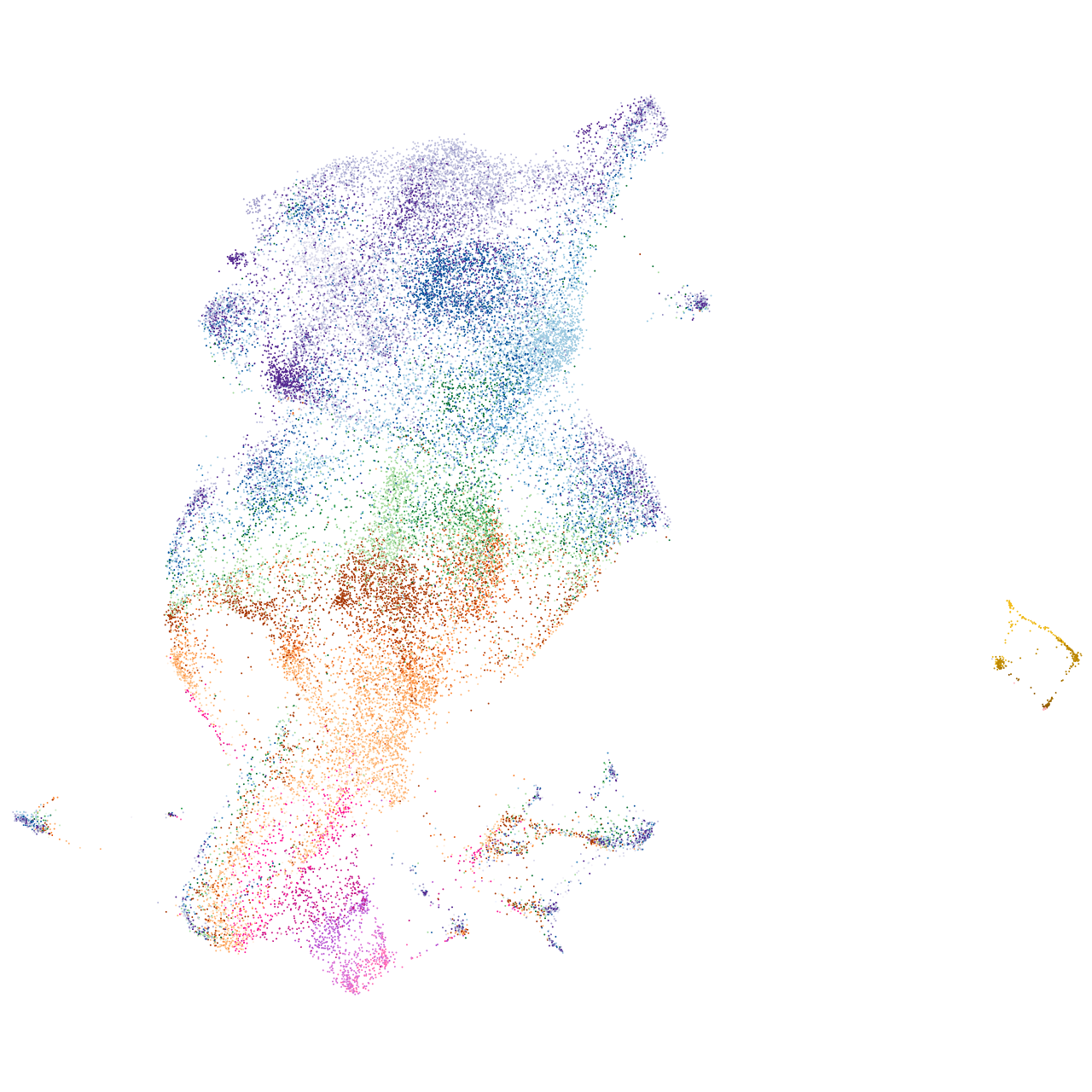prospero homeobox 1a
ZFIN 
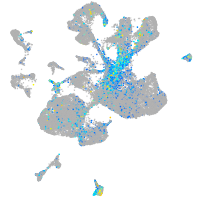
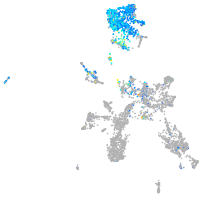
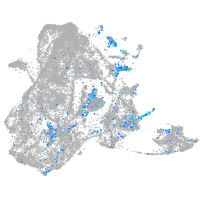
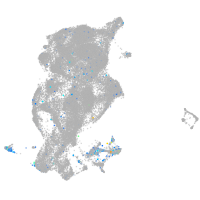
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| cldnh | 0.259 | krt4 | -0.059 |
| grp | 0.249 | cfl1l | -0.054 |
| gfral | 0.231 | cyt1 | -0.051 |
| prr15la | 0.217 | si:ch211-195b11.3 | -0.047 |
| CR759836.1 | 0.217 | cyt1l | -0.042 |
| CABZ01073083.1 | 0.213 | krt5 | -0.042 |
| si:ch211-256e16.6 | 0.203 | wu:fb18f06 | -0.041 |
| pvalb8 | 0.202 | zgc:193505 | -0.040 |
| hepacam2 | 0.198 | agr1 | -0.038 |
| tph1b | 0.180 | zgc:175088 | -0.036 |
| CABZ01095001.1 | 0.179 | krt17 | -0.035 |
| pcbp3 | 0.175 | s100a10b | -0.034 |
| vill | 0.172 | icn2 | -0.032 |
| pcsk2 | 0.171 | krtt1c19e | -0.032 |
| rgs13 | 0.167 | selenow1 | -0.032 |
| cldn8 | 0.161 | zgc:163030 | -0.032 |
| cers3b | 0.158 | si:ch211-157c3.4 | -0.031 |
| ret | 0.156 | mgst1.2 | -0.031 |
| sox1b | 0.155 | cav2 | -0.030 |
| scg3 | 0.153 | pycard | -0.030 |
| mir375-2 | 0.149 | anxa2a | -0.030 |
| neurod4 | 0.148 | tcima | -0.029 |
| zgc:162707 | 0.143 | si:ch73-52f15.5 | -0.029 |
| zgc:101744 | 0.143 | mgst2 | -0.028 |
| dmrta2 | 0.142 | capns1a | -0.028 |
| LOC103911301 | 0.141 | cst14b.1 | -0.027 |
| BX247868.1 | 0.141 | anxa1c | -0.027 |
| mdkb | 0.140 | si:ch211-125o16.4 | -0.027 |
| gpm6aa | 0.140 | si:ch211-105c13.3 | -0.027 |
| ckbb | 0.139 | gsta.1 | -0.027 |
| chga | 0.139 | selenow2b | -0.027 |
| insm1b | 0.139 | si:dkey-184p9.7 | -0.026 |
| LOC110439864 | 0.139 | ahcy | -0.026 |
| hpcal1 | 0.138 | anxa1a | -0.025 |
| gng8 | 0.137 | them6 | -0.025 |