protein kinase cGMP-dependent 1b
ZFIN 
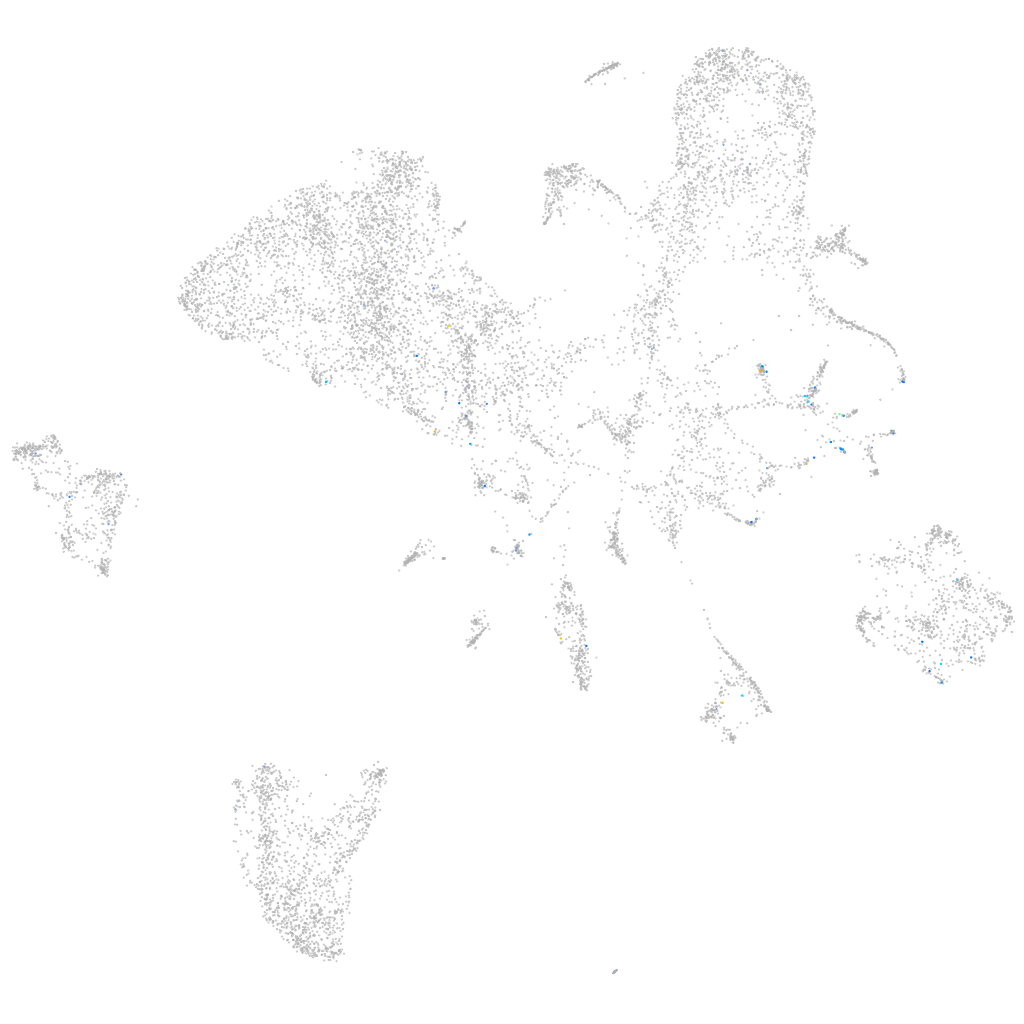
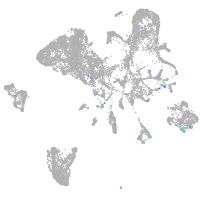
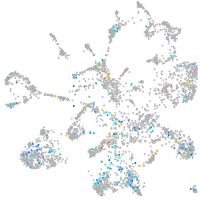
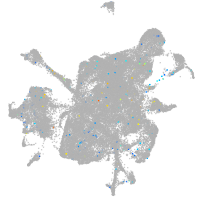
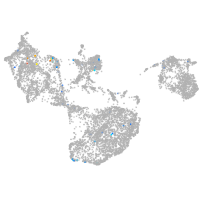
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| FP016239.1 | 0.274 | ahcy | -0.055 |
| tnnc1a | 0.253 | nupr1b | -0.050 |
| LOC110438280 | 0.240 | fbp1b | -0.046 |
| CR847844.2 | 0.231 | gstr | -0.046 |
| nppa | 0.220 | aldob | -0.043 |
| rprml | 0.207 | eno3 | -0.042 |
| si:ch73-208g10.1 | 0.189 | ppa1b | -0.042 |
| LOC108180204 | 0.181 | glud1b | -0.042 |
| gpib | 0.175 | gatm | -0.042 |
| cmlc1 | 0.170 | gapdh | -0.042 |
| ednrab | 0.169 | igf2bp3 | -0.041 |
| obsl1b | 0.165 | cx32.3 | -0.041 |
| AL590146.1 | 0.163 | slc37a4a | -0.039 |
| CU041319.1 | 0.153 | dpydb | -0.039 |
| gcm2 | 0.152 | gamt | -0.039 |
| znf1029 | 0.149 | ugt1a7 | -0.039 |
| BX682558.1 | 0.145 | ppifb | -0.039 |
| BX901974.1 | 0.145 | hspe1 | -0.039 |
| LOC100329490 | 0.143 | sod1 | -0.039 |
| gpr184 | 0.143 | hsdl2 | -0.038 |
| LOC110439392 | 0.142 | suclg1 | -0.038 |
| pde6ga | 0.141 | ndufa6 | -0.038 |
| XLOC-000341 | 0.138 | gstt1a | -0.038 |
| CU862015.1 | 0.134 | suclg2 | -0.038 |
| CR848757.1 | 0.133 | haao | -0.038 |
| LOC101884790 | 0.130 | aldh1l1 | -0.038 |
| CT027815.1 | 0.130 | phb2b | -0.038 |
| LOC101887100 | 0.127 | mat1a | -0.037 |
| ccdc88aa | 0.126 | cox6b1 | -0.037 |
| baalca | 0.124 | pa2g4a | -0.037 |
| BX511010.1 | 0.124 | atp5f1d | -0.036 |
| CU929079.1 | 0.123 | hdlbpa | -0.036 |
| CR774179.5 | 0.122 | fabp3 | -0.036 |
| LOC101884404 | 0.121 | dhrs9 | -0.035 |
| tnni1b | 0.119 | adka | -0.035 |