"protein kinase, cAMP-dependent, catalytic, alpha, genome duplicate a"
ZFIN 
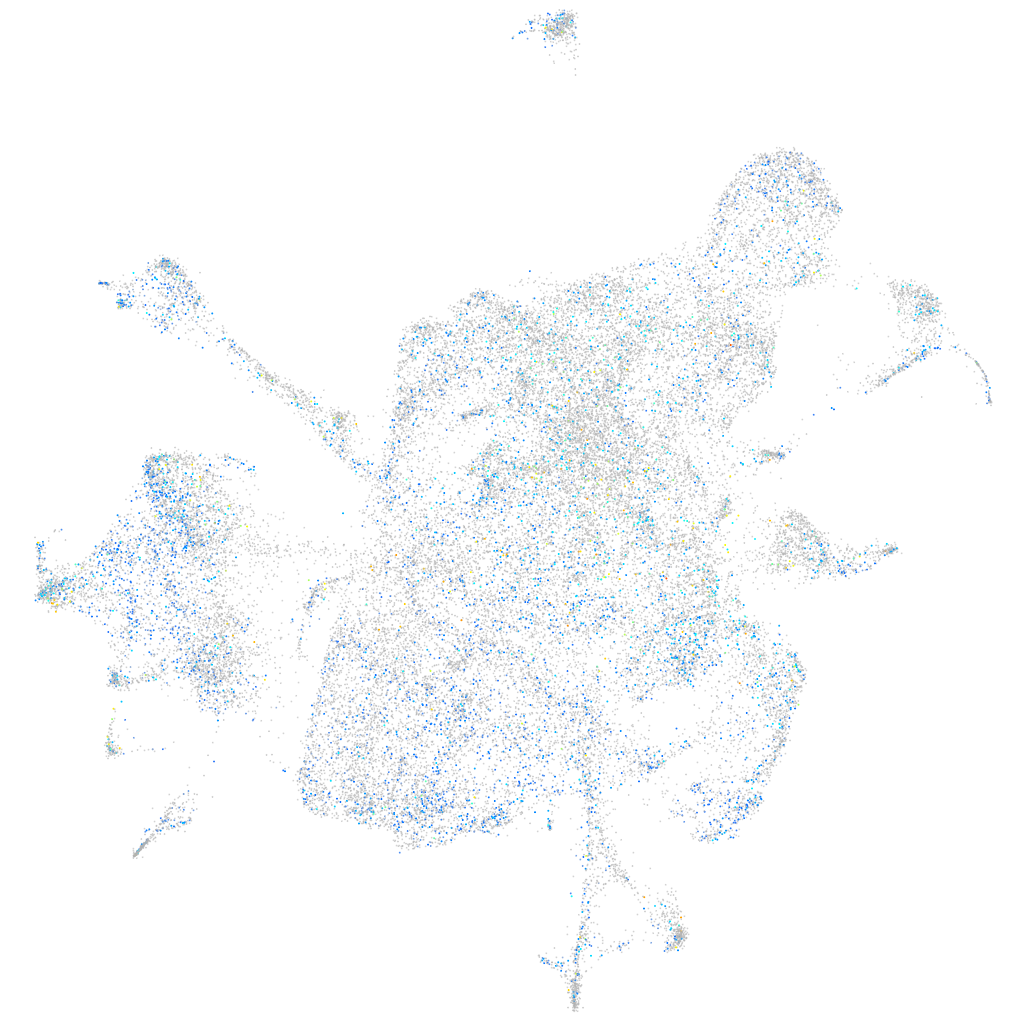
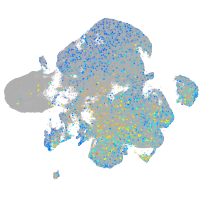
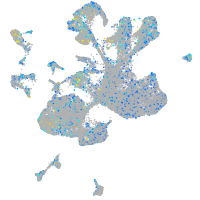
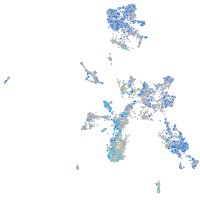
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| marcksl1b | 0.068 | si:dkey-151g10.6 | -0.056 |
| marcksl1a | 0.064 | rplp1 | -0.055 |
| akap12b | 0.063 | rpl39 | -0.055 |
| celf2 | 0.059 | rpl37 | -0.046 |
| gapdhs | 0.058 | rps20 | -0.045 |
| si:ch73-193i22.1 | 0.057 | rpl38 | -0.043 |
| snap25b | 0.057 | rps23 | -0.043 |
| atp1a3a | 0.057 | rps21 | -0.043 |
| CR361564.1 | 0.055 | rpl36a | -0.043 |
| napgb | 0.054 | rpl19 | -0.042 |
| map1aa | 0.053 | rpl26 | -0.042 |
| necap1 | 0.052 | rpl23 | -0.042 |
| kdm6bb | 0.052 | rps25 | -0.041 |
| si:dkey-56m19.5 | 0.052 | rps29 | -0.040 |
| mt-cyb | 0.051 | rps19 | -0.039 |
| tuba1a | 0.051 | rps7 | -0.039 |
| olfm1b | 0.051 | rpl27a | -0.039 |
| atp6v1e1b | 0.050 | rps15a | -0.039 |
| slc4a5a | 0.050 | rps12 | -0.039 |
| zfhx3 | 0.050 | rplp0 | -0.038 |
| tuba1c | 0.050 | rpl35 | -0.037 |
| gng3 | 0.049 | rps13 | -0.037 |
| serinc1 | 0.049 | rplp2l | -0.037 |
| gpm6aa | 0.049 | rps26l | -0.037 |
| nova2 | 0.049 | rpl7 | -0.037 |
| tnr | 0.049 | rps24 | -0.037 |
| vamp2 | 0.049 | faua | -0.036 |
| cotl1 | 0.049 | rpl21 | -0.036 |
| atp6v0cb | 0.049 | rpl10 | -0.035 |
| atp1b2a | 0.049 | rps14 | -0.035 |
| dbn1 | 0.049 | rps2 | -0.035 |
| si:ch211-56a11.2 | 0.049 | rps27.1 | -0.035 |
| pcbp3 | 0.049 | rpl12 | -0.035 |
| BX897741.1 | 0.049 | rpl10a | -0.034 |
| sept15 | 0.049 | rpl29 | -0.034 |