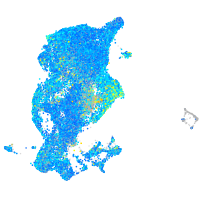
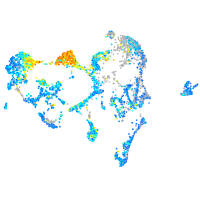
peroxiredoxin 5
ZFIN 
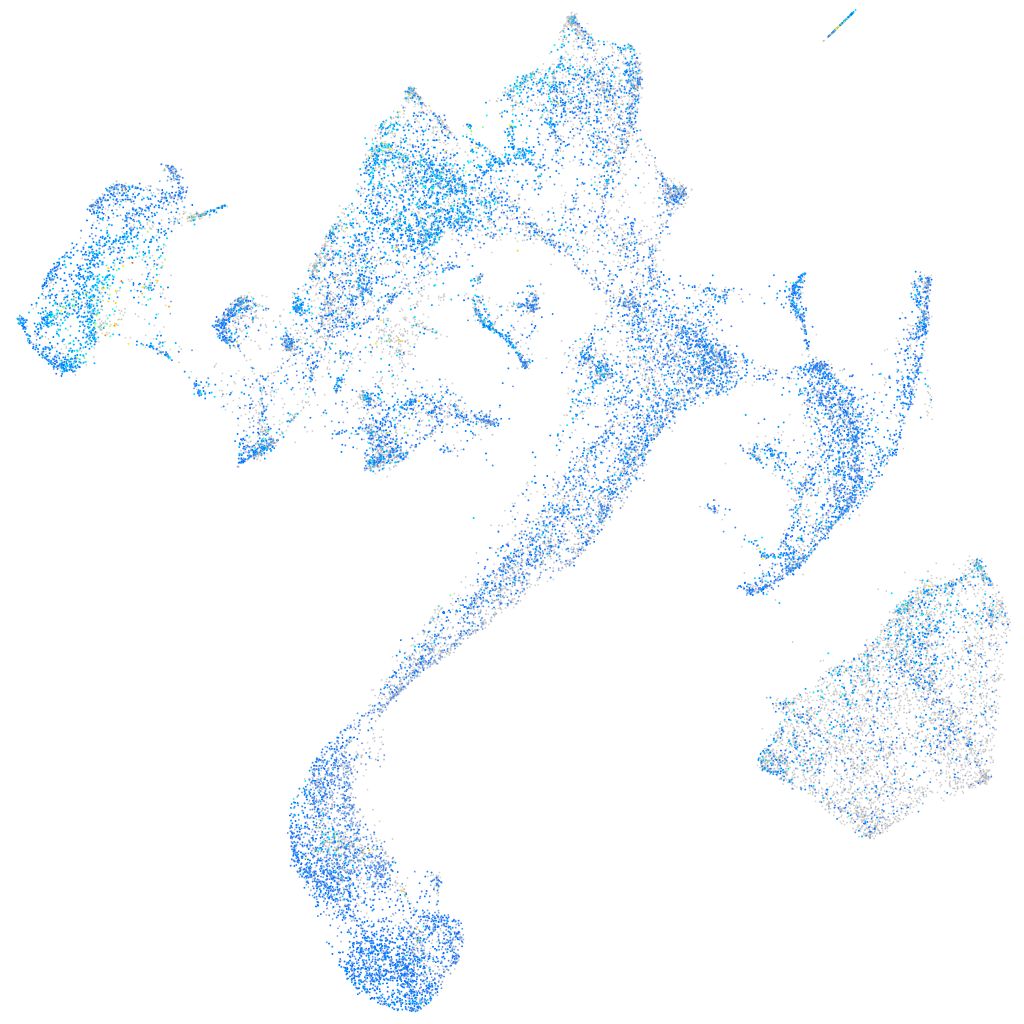
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| atp5mc3b | 0.253 | stm | -0.205 |
| atp5f1b | 0.252 | NC-002333.4 | -0.204 |
| atp5mc1 | 0.242 | hnrnpa1a | -0.203 |
| atp5meb | 0.238 | ncl | -0.199 |
| atp5l | 0.236 | hnrnpub | -0.198 |
| idh2 | 0.235 | hnrnpm | -0.194 |
| atp5fa1 | 0.230 | hnrnpa1b | -0.192 |
| cox4i1 | 0.226 | anp32e | -0.188 |
| vdac2 | 0.224 | hspb1 | -0.188 |
| ndufb10 | 0.223 | top1l | -0.187 |
| mdh2 | 0.223 | marcksb | -0.186 |
| cox8a | 0.222 | rbm4.3 | -0.186 |
| cox6a1 | 0.221 | srrm1 | -0.185 |
| atp5pd | 0.221 | ppig | -0.184 |
| sod2 | 0.219 | dkc1 | -0.183 |
| atp5pf | 0.217 | nucks1a | -0.183 |
| ldhba | 0.215 | akap12b | -0.181 |
| ndufb7 | 0.213 | si:dkey-56m19.5 | -0.180 |
| atp5f1c | 0.213 | snrnp70 | -0.180 |
| ndufa3 | 0.211 | fbl | -0.180 |
| vdac3 | 0.211 | syncrip | -0.180 |
| pvalb4 | 0.208 | nop56 | -0.179 |
| mpc1 | 0.208 | ilf3b | -0.178 |
| ndufb3 | 0.206 | nop58 | -0.178 |
| suclg1 | 0.205 | BX927327.1 | -0.176 |
| cox7a2a | 0.204 | acin1a | -0.176 |
| cox5ab | 0.203 | acin1b | -0.176 |
| atp5f1d | 0.203 | brd3a | -0.175 |
| ndufb8 | 0.202 | srrm2 | -0.175 |
| mt-nd1 | 0.201 | npm1a | -0.174 |
| mdh1aa | 0.200 | thrap3b | -0.174 |
| atp5if1b | 0.200 | arf1 | -0.174 |
| COX5B | 0.200 | pabpc1a | -0.173 |
| atp5pb | 0.200 | pou5f3 | -0.173 |
| mt-atp6 | 0.200 | bms1 | -0.172 |