PPARG related coactivator 1
ZFIN 
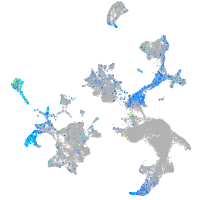
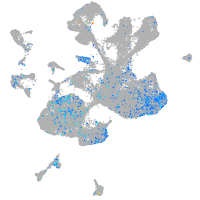
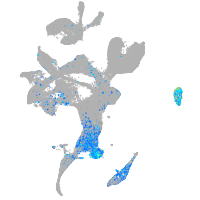
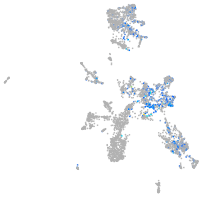
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| dkc1 | 0.559 | rpl37 | -0.568 |
| nop58 | 0.555 | rps10 | -0.553 |
| nop2 | 0.552 | zgc:114188 | -0.523 |
| nop56 | 0.537 | rps17 | -0.474 |
| zmp:0000000624 | 0.537 | atp1b1a | -0.415 |
| apoc1 | 0.534 | atp5f1b | -0.404 |
| ncl | 0.533 | atp5mc1 | -0.403 |
| npm1a | 0.533 | zgc:56493 | -0.402 |
| fbl | 0.510 | atp5mc3b | -0.398 |
| cdx4 | 0.505 | atp5l | -0.382 |
| hspb1 | 0.500 | slc25a5 | -0.381 |
| rsl1d1 | 0.499 | atp5fa1 | -0.367 |
| wu:fb97g03 | 0.498 | vdac3 | -0.366 |
| rrp1 | 0.496 | cox5aa | -0.365 |
| mybbp1a | 0.488 | cdh17 | -0.350 |
| gnl3 | 0.488 | mdh1aa | -0.349 |
| ppig | 0.479 | suclg1 | -0.347 |
| apoeb | 0.478 | atp5meb | -0.342 |
| NC-002333.4 | 0.469 | ndrg1a | -0.340 |
| mphosph10 | 0.466 | ndufa4l | -0.340 |
| bms1 | 0.466 | eno3 | -0.339 |
| pes | 0.465 | hnrnpa0l | -0.336 |
| ebna1bp2 | 0.463 | slc25a3b | -0.333 |
| lyar | 0.463 | COX5B | -0.332 |
| snrnp70 | 0.455 | mt-nd1 | -0.332 |
| gar1 | 0.451 | atp5f1d | -0.331 |
| rbmx2 | 0.450 | h3f3a | -0.329 |
| anp32e | 0.449 | atp5if1b | -0.326 |
| lig1 | 0.447 | atp5pf | -0.326 |
| rrp15 | 0.445 | cox8a | -0.325 |
| anp32a | 0.442 | vdac2 | -0.325 |
| hnrnpub | 0.438 | mdh2 | -0.324 |
| acin1a | 0.437 | si:ch211-139a5.9 | -0.323 |
| pinx1 | 0.432 | cox6a1 | -0.321 |
| hnrnpa1b | 0.430 | tpi1b | -0.319 |