"protein phosphatase 1, regulatory subunit 42"
ZFIN 
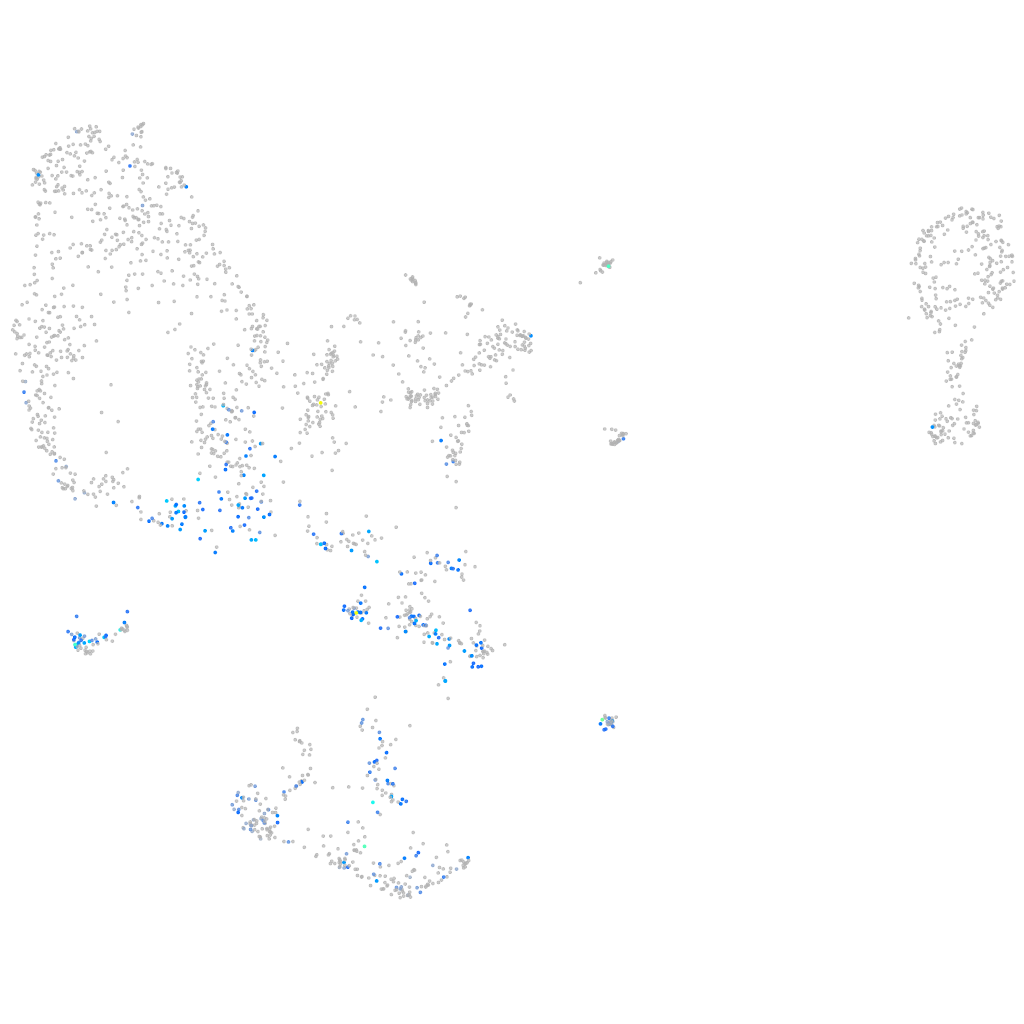
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros 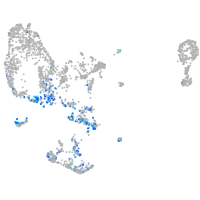 |
neural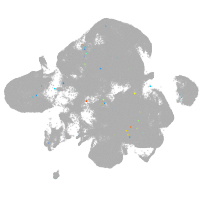 |
spinal cord / glia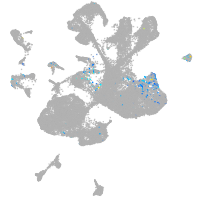 |
eye |
taste / olfactory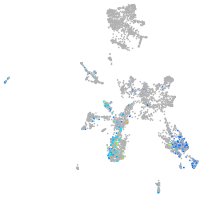 |
otic / lateral line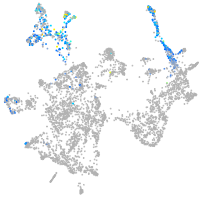 |
epidermis |
periderm |
fin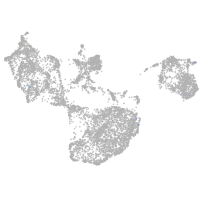 |
ionocytes / mucous |
pigment cells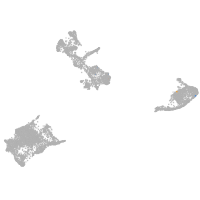 |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| daw1 | 0.374 | phyhd1 | -0.177 |
| tctex1d1 | 0.372 | gcshb | -0.176 |
| cfap45 | 0.368 | gapdh | -0.173 |
| ccdc151 | 0.365 | agxtb | -0.170 |
| si:ch211-163l21.7 | 0.362 | alpl | -0.170 |
| foxj1a | 0.361 | si:dkey-33i11.9 | -0.166 |
| spag6 | 0.360 | cx32.3 | -0.164 |
| efhc2 | 0.356 | epdl2 | -0.164 |
| capsla | 0.353 | nipsnap3a | -0.160 |
| RSPH1 | 0.353 | haao | -0.159 |
| spata4 | 0.352 | gstt1a | -0.156 |
| ccdc146 | 0.347 | rgn | -0.156 |
| dnali1 | 0.344 | g6pca.2 | -0.153 |
| ribc2 | 0.343 | slc20a1a | -0.152 |
| pacrg | 0.334 | gpt2l | -0.151 |
| rsph9 | 0.334 | ctsz | -0.151 |
| tekt4 | 0.333 | dpys | -0.150 |
| mycbp | 0.329 | slc5a12 | -0.149 |
| enkur | 0.328 | slc26a6l | -0.149 |
| LOC103911624 | 0.327 | dhrs9 | -0.148 |
| efhc1 | 0.327 | slc16a1b | -0.148 |
| ccdc173 | 0.324 | si:ch211-201h21.5 | -0.146 |
| si:dkey-114c15.5 | 0.323 | slc22a13b | -0.145 |
| si:ch211-248e11.2 | 0.321 | slc5a11 | -0.145 |
| cfap20 | 0.321 | pnp6 | -0.142 |
| catip | 0.320 | acy1 | -0.142 |
| meig1 | 0.317 | slc34a1a | -0.142 |
| iqcg | 0.315 | ASS1 | -0.141 |
| capslb | 0.314 | akr7a3 | -0.141 |
| spaca9 | 0.313 | pvalb9 | -0.139 |
| cldnc | 0.312 | slc15a2 | -0.138 |
| dnal1 | 0.311 | tspan36 | -0.138 |
| ropn1l | 0.311 | slc13a3 | -0.137 |
| ttc25 | 0.310 | CR381686.5 | -0.137 |
| nme5 | 0.310 | etnppl | -0.136 |