"protein phosphatase 1, regulatory subunit 3Ca"
ZFIN 
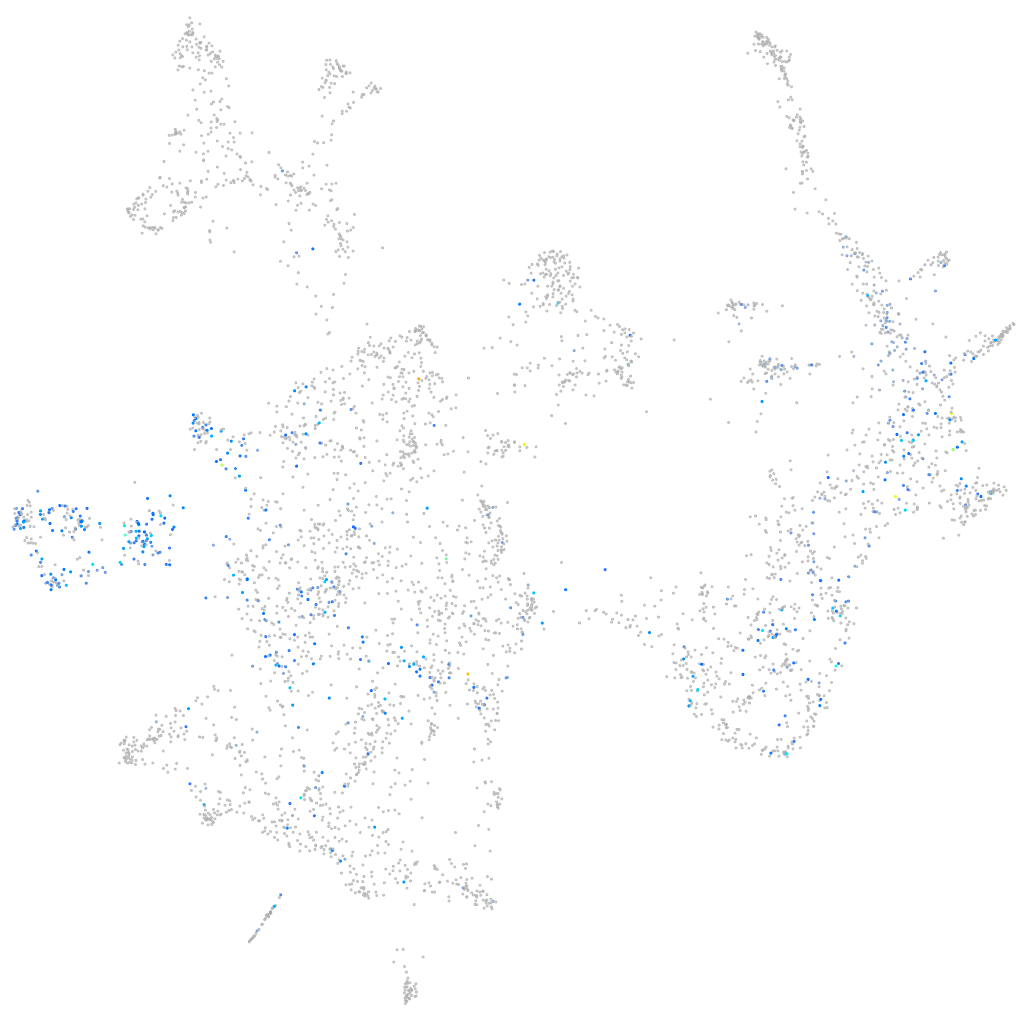
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| zgc:194210 | 0.269 | COX3 | -0.172 |
| CABZ01075068.1 | 0.252 | pvalb1 | -0.156 |
| zbtb16a | 0.249 | mt-atp6 | -0.142 |
| wu:fc01d11 | 0.240 | pvalb2 | -0.142 |
| s1pr5a | 0.234 | atp2b1a | -0.136 |
| oc90 | 0.232 | mt-co2 | -0.131 |
| dnase1l4.1 | 0.221 | actc1b | -0.125 |
| LOC100331480 | 0.221 | gapdhs | -0.123 |
| eif4ebp3l | 0.219 | ckbb | -0.122 |
| ppp1r3b | 0.216 | calm1b | -0.118 |
| npm1a | 0.213 | cox7c | -0.116 |
| eef1a1l1 | 0.208 | nptna | -0.113 |
| cited4b | 0.207 | zgc:158463 | -0.113 |
| hnrnpa0l | 0.206 | otofb | -0.111 |
| sox9b | 0.204 | sod2 | -0.111 |
| ednrab | 0.204 | eno1a | -0.111 |
| fam20cb | 0.204 | col9a1a | -0.110 |
| zfp36l1a | 0.202 | atp1a3b | -0.110 |
| khdrbs1a | 0.198 | cd164l2 | -0.110 |
| lin28a | 0.195 | tpi1a | -0.109 |
| prdm12b | 0.195 | b2ml | -0.109 |
| mycn | 0.191 | gpx2 | -0.108 |
| nop58 | 0.190 | dnajc5b | -0.107 |
| ccnd1 | 0.189 | osbpl1a | -0.106 |
| sp7 | 0.188 | atp6v0cb | -0.106 |
| si:ch211-222l21.1 | 0.188 | kif1aa | -0.105 |
| cldna | 0.186 | atp5mc1 | -0.104 |
| dlx3b | 0.186 | atp6v1e1b | -0.104 |
| marcksl1b | 0.186 | mt-nd4 | -0.103 |
| nhp2 | 0.184 | lrrc73 | -0.103 |
| scn4bb | 0.182 | calml4a | -0.103 |
| zbtb16b | 0.182 | emb | -0.102 |
| nop56 | 0.181 | CR925719.1 | -0.102 |
| apex1 | 0.180 | si:ch73-15n15.3 | -0.102 |
| akt1s1 | 0.178 | atp5mf | -0.102 |