"protein phosphatase, Mg2+/Mn2+ dependent, 1Da"
ZFIN 
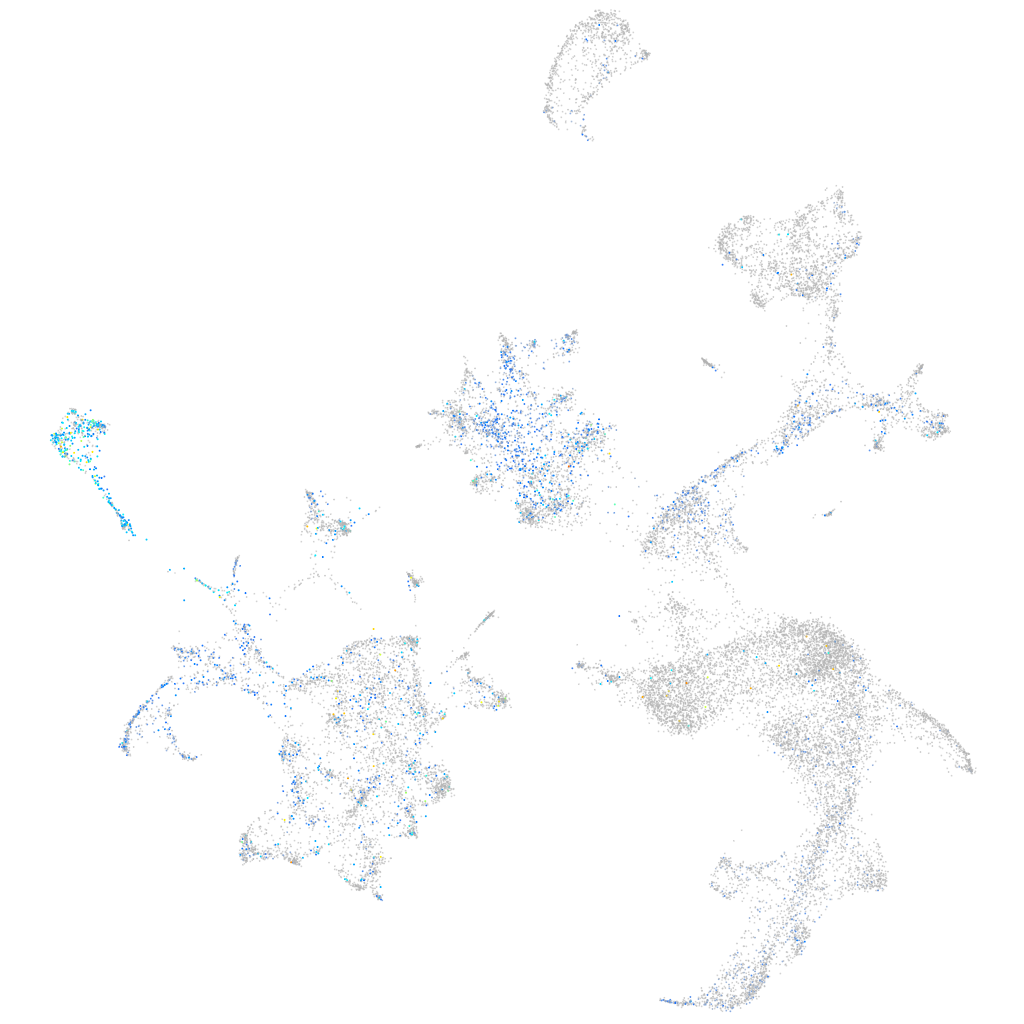
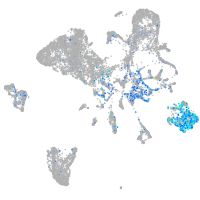
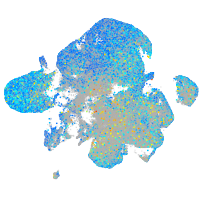
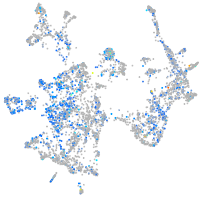
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| cx43.4 | 0.326 | hbbe1.3 | -0.136 |
| marcksb | 0.317 | hbae3 | -0.134 |
| hspb1 | 0.316 | prdx2 | -0.134 |
| hnrnpa1b | 0.292 | hbae1.1 | -0.133 |
| ppig | 0.290 | hbae1.3 | -0.129 |
| stm | 0.290 | hbbe2 | -0.127 |
| hnrnpub | 0.288 | hbbe1.1 | -0.127 |
| pou5f3 | 0.282 | cahz | -0.123 |
| nucks1a | 0.282 | hemgn | -0.120 |
| ilf3b | 0.280 | blvrb | -0.118 |
| snrnp70 | 0.277 | hbbe1.2 | -0.114 |
| si:ch73-281n10.2 | 0.277 | alas2 | -0.114 |
| si:ch211-137a8.4 | 0.276 | gpx1a | -0.113 |
| hmga1a | 0.276 | nt5c2l1 | -0.113 |
| si:ch211-288g17.3 | 0.274 | tspo | -0.112 |
| seta | 0.267 | creg1 | -0.109 |
| hnrnpa0b | 0.266 | mcl1a | -0.108 |
| ncl | 0.265 | zgc:163057 | -0.104 |
| zic2b | 0.264 | slc4a1a | -0.103 |
| syncrip | 0.263 | si:ch211-207c6.2 | -0.096 |
| lig1 | 0.263 | plac8l1 | -0.095 |
| COX7A2 | 0.263 | tmod4 | -0.095 |
| hnrnpabb | 0.262 | epb41b | -0.092 |
| anp32e | 0.261 | si:ch211-250g4.3 | -0.092 |
| rbm4.3 | 0.261 | nmt1b | -0.087 |
| khdrbs1a | 0.260 | pvalb1 | -0.085 |
| NC-002333.4 | 0.259 | pvalb2 | -0.084 |
| hnrnpaba | 0.258 | actc1b | -0.084 |
| brd3a | 0.258 | selenoj | -0.083 |
| sp5l | 0.257 | vdac2 | -0.080 |
| hnrnpa1a | 0.257 | hbae5 | -0.078 |
| si:ch211-152c2.3 | 0.256 | si:ch211-227m13.1 | -0.077 |
| si:ch73-1a9.3 | 0.254 | bloc1s6 | -0.076 |
| srsf1a | 0.253 | hif1al2 | -0.073 |
| ppm1g | 0.252 | gabarapa | -0.072 |