peptidylprolyl isomerase Fb
ZFIN 
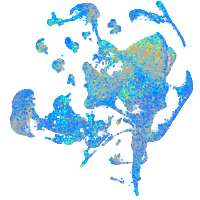
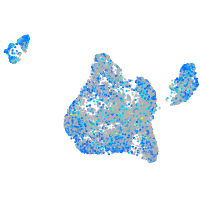
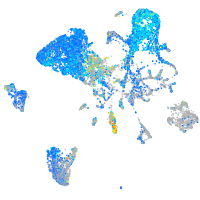
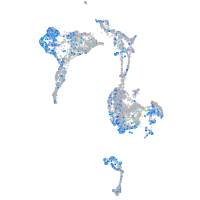
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| suclg1 | 0.554 | hspb1 | -0.469 |
| eno3 | 0.546 | marcksb | -0.452 |
| ldhba | 0.544 | wu:fb97g03 | -0.441 |
| atp5mc3b | 0.539 | si:ch73-1a9.3 | -0.440 |
| atp5mc1 | 0.534 | nucks1a | -0.438 |
| atp5f1b | 0.531 | hmgb2b | -0.429 |
| mdh1aa | 0.516 | si:ch73-281n10.2 | -0.427 |
| atp5pd | 0.515 | hmga1a | -0.420 |
| atp5meb | 0.504 | seta | -0.420 |
| atp5fa1 | 0.502 | hnrnpabb | -0.410 |
| atp5f1d | 0.502 | apoeb | -0.403 |
| tpi1b | 0.498 | marcksl1b | -0.400 |
| si:ch211-139a5.9 | 0.494 | hnrnpaba | -0.398 |
| atp5l | 0.491 | h2afvb | -0.390 |
| atp1b1a | 0.490 | hmgb2a | -0.388 |
| glud1b | 0.484 | cx43.4 | -0.383 |
| atp6v1e1b | 0.480 | hmgb1b | -0.382 |
| atp5pb | 0.479 | ilf3b | -0.378 |
| atp5if1b | 0.479 | setb | -0.374 |
| cox7a2a | 0.478 | hmgn2 | -0.371 |
| prdx3 | 0.478 | hmgn6 | -0.370 |
| gstp1 | 0.478 | apoc1 | -0.368 |
| ndufa4l | 0.475 | syncrip | -0.367 |
| cdh17 | 0.475 | si:ch211-288g17.3 | -0.365 |
| cox5ab | 0.472 | ptmab | -0.361 |
| aldh6a1 | 0.472 | hnrnpa1a | -0.361 |
| cox6a1 | 0.470 | anp32a | -0.356 |
| COX5B | 0.470 | h3f3d | -0.355 |
| atp5f1c | 0.469 | anp32b | -0.355 |
| gcshb | 0.468 | hnrnpub | -0.353 |
| sod1 | 0.467 | cirbpa | -0.353 |
| cox6b1 | 0.465 | acin1a | -0.353 |
| ndufb8 | 0.464 | msna | -0.346 |
| atp5pf | 0.461 | akap12b | -0.346 |
| sod2 | 0.460 | ncl | -0.343 |