periphilin 1
ZFIN 
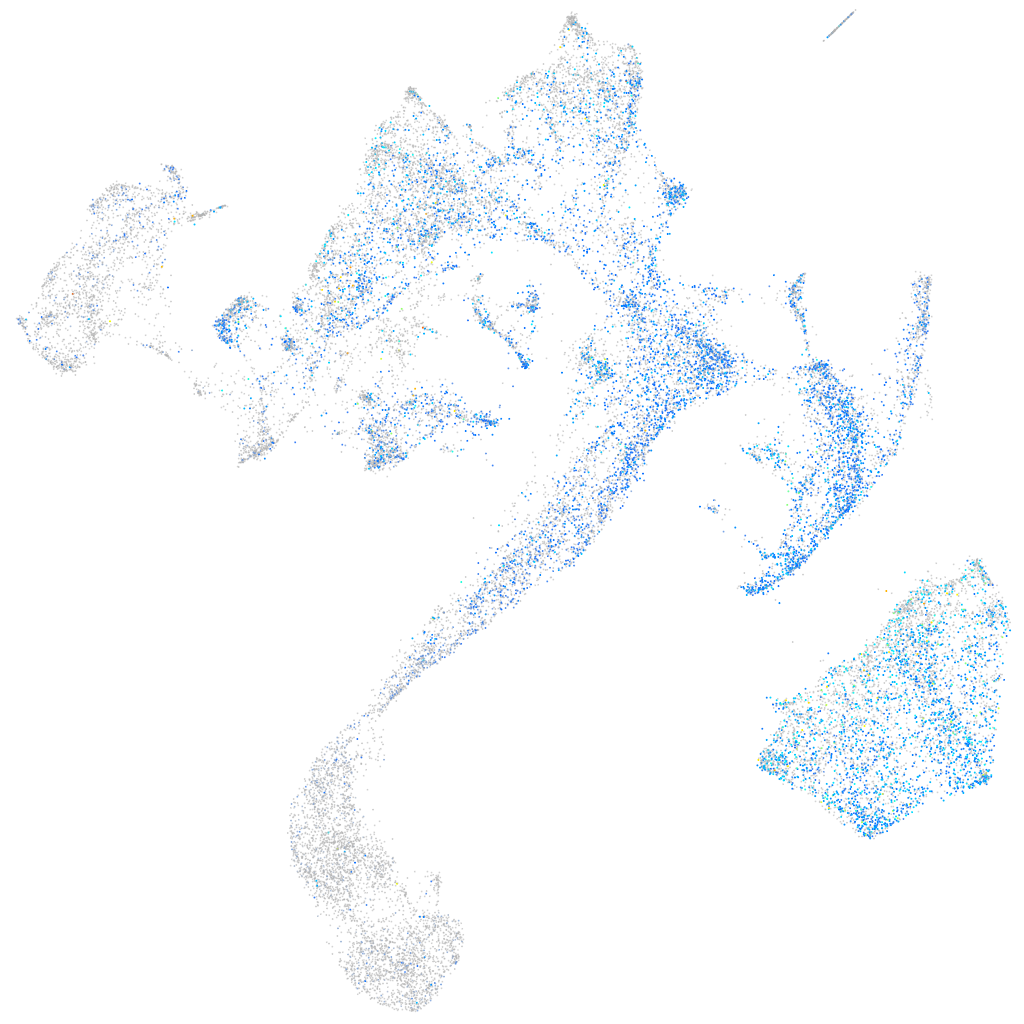
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| hnrnpabb | 0.325 | actc1b | -0.276 |
| khdrbs1a | 0.318 | ckmb | -0.256 |
| hmga1a | 0.315 | ckma | -0.256 |
| hnrnpaba | 0.310 | ak1 | -0.255 |
| hnrnpa0b | 0.308 | atp2a1 | -0.251 |
| setb | 0.307 | tnnc2 | -0.247 |
| anp32b | 0.306 | aldoab | -0.244 |
| cbx3a | 0.305 | neb | -0.243 |
| hmgb2a | 0.304 | ttn.2 | -0.235 |
| h2afvb | 0.300 | ldb3b | -0.233 |
| cirbpa | 0.297 | acta1b | -0.233 |
| hmgb2b | 0.296 | mylpfa | -0.232 |
| seta | 0.295 | si:ch73-367p23.2 | -0.232 |
| cx43.4 | 0.294 | nme2b.2 | -0.231 |
| dkc1 | 0.291 | actn3a | -0.231 |
| snrpb | 0.291 | eno3 | -0.230 |
| ncl | 0.289 | tmem38a | -0.230 |
| nop58 | 0.289 | actn3b | -0.229 |
| rbm8a | 0.289 | myom1a | -0.227 |
| nop56 | 0.289 | gapdh | -0.227 |
| npm1a | 0.288 | pvalb1 | -0.227 |
| syncrip | 0.288 | tpma | -0.227 |
| nucks1a | 0.286 | ttn.1 | -0.225 |
| si:ch211-222l21.1 | 0.286 | ldb3a | -0.225 |
| si:ch73-281n10.2 | 0.286 | smyd1a | -0.225 |
| hdac1 | 0.285 | pabpc4 | -0.224 |
| srsf3b | 0.285 | pvalb2 | -0.223 |
| cirbpb | 0.284 | mylz3 | -0.223 |
| h3f3d | 0.281 | tmod4 | -0.222 |
| sumo3a | 0.281 | tnnt3a | -0.222 |
| ddx39ab | 0.281 | si:ch211-266g18.10 | -0.222 |
| srsf2a | 0.280 | pgam2 | -0.222 |
| hmgn7 | 0.278 | CABZ01078594.1 | -0.220 |
| snrpd1 | 0.278 | ank1a | -0.220 |
| snrpf | 0.276 | eno1a | -0.218 |