pancreatic progenitor cell differentiation and proliferation factor b
ZFIN 
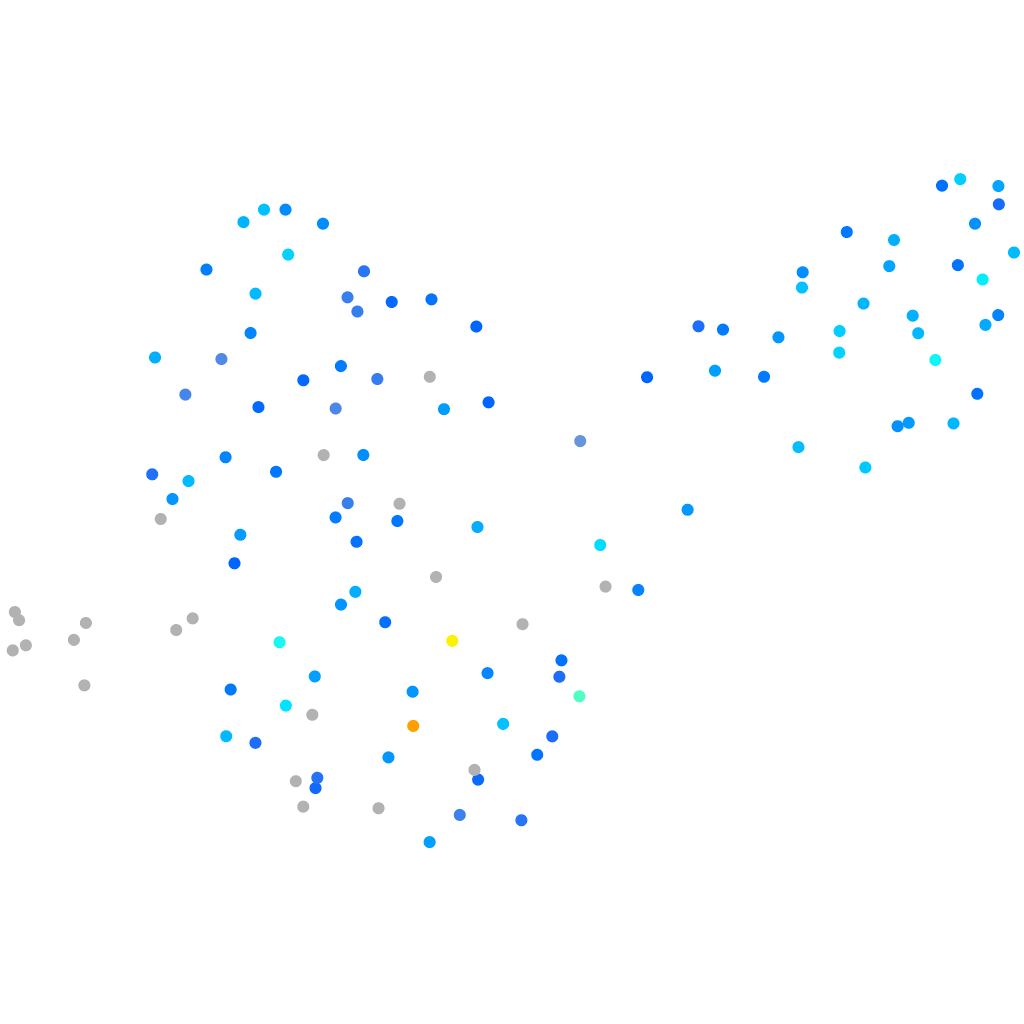
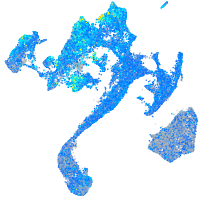
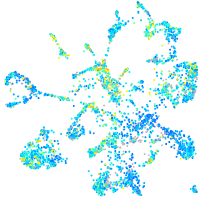
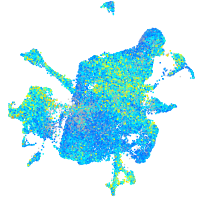
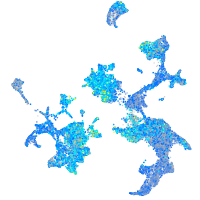
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| dap1b | 0.524 | NC-002333.4 | -0.496 |
| srd5a2a | 0.505 | marcksb | -0.422 |
| gabarapb | 0.496 | snrpa | -0.410 |
| vdac2 | 0.479 | hnrnpub | -0.409 |
| rps26l | 0.475 | nop58 | -0.407 |
| h3f3a | 0.465 | npm1a | -0.399 |
| mt-nd1 | 0.465 | syncrip | -0.391 |
| cox6a1 | 0.464 | ccdc9 | -0.378 |
| zgc:92335 | 0.464 | ccna2 | -0.361 |
| zgc:92744 | 0.464 | smc1al | -0.359 |
| rps13 | 0.460 | snrpb2 | -0.353 |
| btf3 | 0.460 | syncripl | -0.352 |
| ccni | 0.456 | tcea1 | -0.351 |
| mgst3b | 0.452 | fscn1a | -0.351 |
| ISCU (1 of many) | 0.450 | hdgfl2 | -0.350 |
| atp6v1e1b | 0.449 | zc3h13 | -0.347 |
| txnipa | 0.447 | lig1 | -0.347 |
| gsta.1 | 0.437 | smarcd1 | -0.343 |
| tubb4b | 0.436 | pnn | -0.343 |
| sec61g | 0.434 | utp3 | -0.343 |
| zgc:158463 | 0.433 | zmat2 | -0.342 |
| naca | 0.430 | hnrnpa1b | -0.339 |
| dpp7 | 0.429 | fbl | -0.337 |
| hsp90ab1 | 0.429 | larp7 | -0.337 |
| rps10 | 0.427 | eloa | -0.336 |
| rpl8 | 0.424 | ppig | -0.336 |
| rpl37 | 0.423 | gle1 | -0.335 |
| ppiab | 0.422 | exd2 | -0.335 |
| lrrc17 | 0.421 | CABZ01044053.1 | -0.335 |
| ndufb9 | 0.421 | srrm1 | -0.332 |
| trappc1 | 0.419 | CT030188.1 | -0.327 |
| cox8a | 0.418 | ap2s1 | -0.325 |
| ube2v1 | 0.418 | dhx38 | -0.325 |
| rpl24 | 0.418 | ncl | -0.324 |
| gng7 | 0.417 | depdc1a | -0.323 |