POU class 3 homeobox 1
ZFIN 
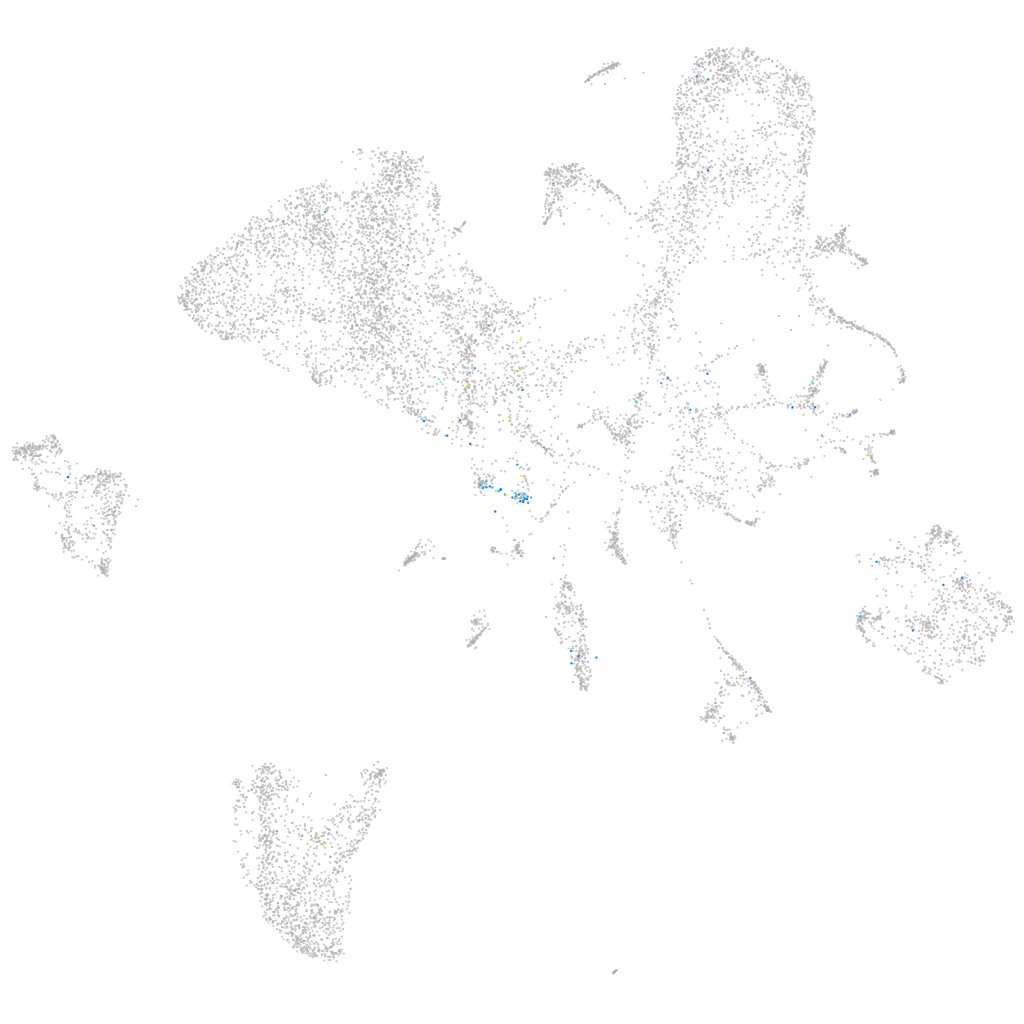
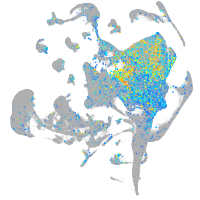
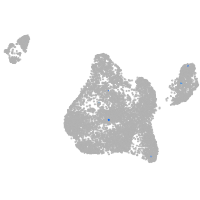
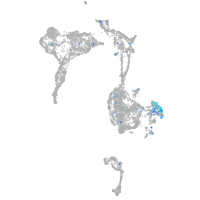
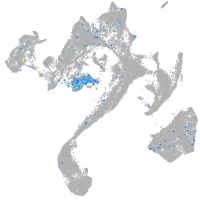
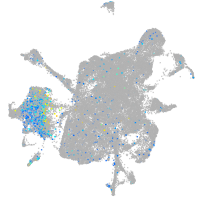
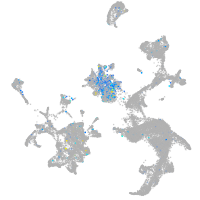
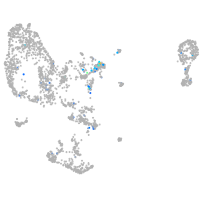
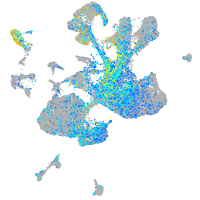
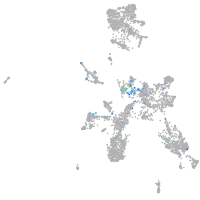
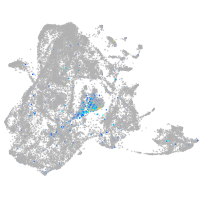
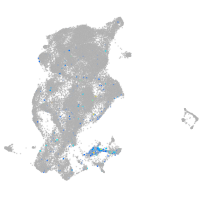
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| LOC798783 | 0.249 | eef1da | -0.081 |
| tuba1c | 0.246 | eno3 | -0.072 |
| ebf2 | 0.242 | aldob | -0.066 |
| elavl3 | 0.242 | cebpd | -0.064 |
| CU861477.1 | 0.237 | hdlbpa | -0.064 |
| LOC101882472 | 0.230 | gapdh | -0.064 |
| scrt2 | 0.230 | hadh | -0.057 |
| neurog1 | 0.230 | cx32.3 | -0.057 |
| XLOC-003689 | 0.228 | suclg1 | -0.056 |
| CU634008.1 | 0.223 | BX908782.3 | -0.056 |
| abhd6a | 0.219 | pklr | -0.056 |
| XLOC-003690 | 0.219 | aldh7a1 | -0.056 |
| XLOC-042919 | 0.217 | haao | -0.055 |
| msi1 | 0.217 | scp2a | -0.055 |
| si:ch211-57n23.4 | 0.215 | aldh9a1a.1 | -0.055 |
| inavaa | 0.215 | tuba8l2 | -0.055 |
| gadd45gb.1 | 0.215 | sod1 | -0.055 |
| nova2 | 0.211 | abat | -0.055 |
| tuba1a | 0.205 | fdx1 | -0.055 |
| BX530077.1 | 0.201 | gstt1a | -0.055 |
| si:ch73-21g5.7 | 0.200 | gpx1a | -0.054 |
| ncam1a | 0.199 | srd5a2a | -0.054 |
| barhl1b | 0.199 | gstk1 | -0.053 |
| sulf2a | 0.197 | cox7a1 | -0.053 |
| plp1a | 0.192 | prdx6 | -0.053 |
| CU467822.1 | 0.192 | dap | -0.053 |
| prob1 | 0.191 | gstr | -0.052 |
| pax7b | 0.191 | sec61b | -0.052 |
| gpm6bb | 0.189 | gstp1 | -0.052 |
| tubb5 | 0.188 | pgk1 | -0.052 |
| scrt1a | 0.187 | atp5meb | -0.052 |
| zgc:165461 | 0.186 | rmdn1 | -0.052 |
| nhlh2 | 0.183 | acadm | -0.051 |
| XLOC-003692 | 0.183 | gamt | -0.051 |
| LOC100007824 | 0.181 | nipsnap3a | -0.051 |