"PMS1 homolog 2, mismatch repair system component"
ZFIN 
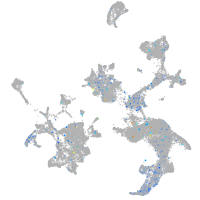
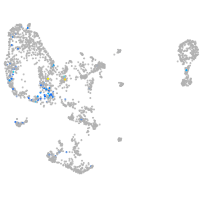
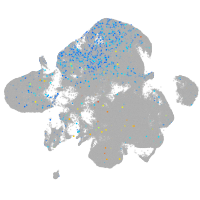
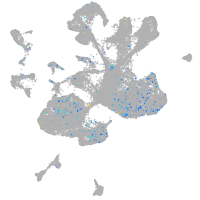
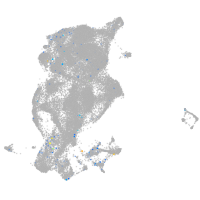
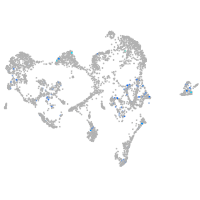
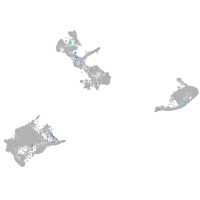
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| si:ch211-8c17.2 | 0.208 | apoa4b.1 | -0.061 |
| CR354612.1 | 0.188 | pnp4b | -0.060 |
| LOC101884197 | 0.181 | etnppl | -0.058 |
| CR405682.1 | 0.170 | gpx4a | -0.057 |
| si:dkey-47k20.5 | 0.168 | fabp10a | -0.057 |
| sspn | 0.164 | apoc1 | -0.057 |
| BX005024.2 | 0.159 | apoa1b | -0.056 |
| LOC101882015 | 0.152 | apoa2 | -0.056 |
| mcm7 | 0.144 | zgc:123103 | -0.056 |
| wisp2 | 0.143 | c9 | -0.056 |
| slc17a8 | 0.142 | si:dkey-86l18.10 | -0.056 |
| zmp:0000000932 | 0.141 | scp2a | -0.055 |
| mcm4 | 0.140 | LOC110437731 | -0.055 |
| znf975 | 0.139 | mgst1.2 | -0.054 |
| si:ch73-190f9.4 | 0.136 | ttr | -0.054 |
| rasal1b | 0.134 | gc | -0.054 |
| XLOC-016403 | 0.132 | ces2 | -0.054 |
| arg1 | 0.132 | apom | -0.053 |
| CR749168.4 | 0.127 | abat | -0.053 |
| LOC100332443 | 0.127 | serpina1 | -0.053 |
| smtnl1 | 0.125 | gatm | -0.053 |
| gins2 | 0.122 | fgb | -0.053 |
| smtna | 0.121 | agxtb | -0.052 |
| pcna | 0.120 | apoc2 | -0.052 |
| LOC101885065 | 0.120 | fga | -0.052 |
| mcm5 | 0.119 | cyp2ad2 | -0.052 |
| caspbl | 0.119 | serpina1l | -0.052 |
| mcm2 | 0.118 | afp4 | -0.052 |
| rrm2 | 0.118 | ugp2a | -0.051 |
| slbp | 0.117 | ambp | -0.051 |
| LOC564610 | 0.115 | fetub | -0.051 |
| dnmt1 | 0.114 | cyp4v8 | -0.051 |
| igfn1.1 | 0.114 | rbp4 | -0.051 |
| asf1ba | 0.113 | dhrs9 | -0.051 |
| msh6 | 0.112 | gda | -0.051 |