plexin domain containing 2
ZFIN 
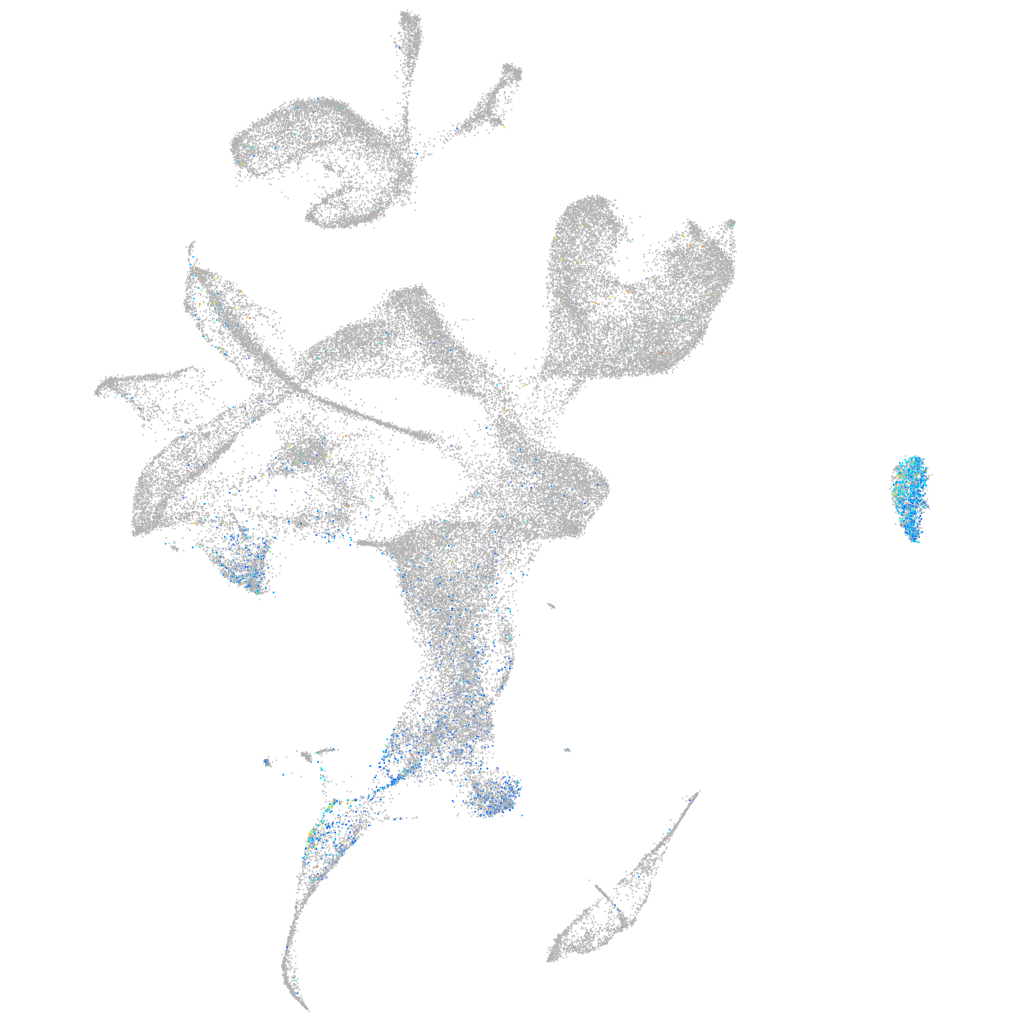
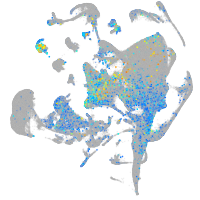
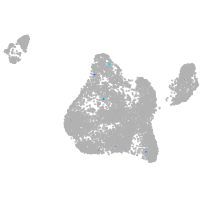
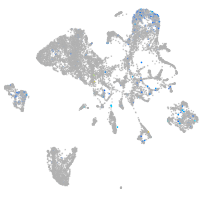
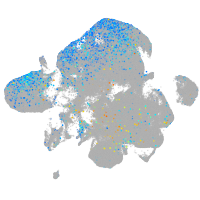
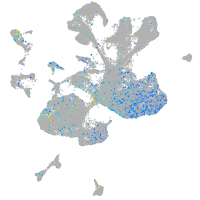
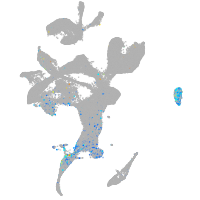
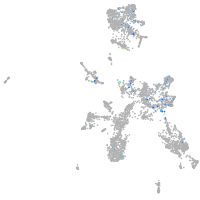
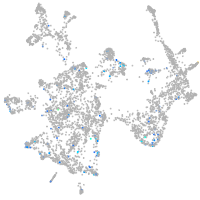
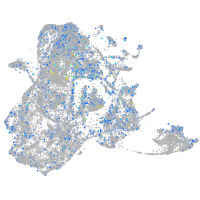
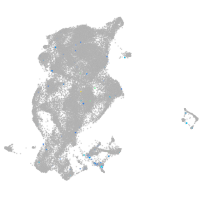
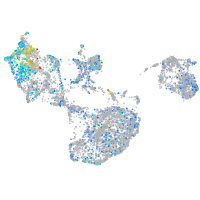
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| hspb1 | 0.354 | ptmaa | -0.157 |
| shisa2a | 0.329 | rpl37 | -0.156 |
| cyp26a1 | 0.324 | rps10 | -0.151 |
| stm | 0.320 | ppiab | -0.149 |
| tdgf1 | 0.310 | h3f3a | -0.149 |
| sox19a | 0.309 | ckbb | -0.144 |
| si:ch211-152c2.3 | 0.307 | si:ch1073-429i10.3.1 | -0.135 |
| apoeb | 0.302 | zgc:114188 | -0.132 |
| nr6a1a | 0.284 | CR383676.1 | -0.132 |
| aldob | 0.284 | tuba1c | -0.126 |
| hesx1 | 0.281 | hnrnpa0l | -0.122 |
| chrd | 0.272 | gpm6aa | -0.122 |
| zmp:0000000624 | 0.266 | gpm6ab | -0.115 |
| NC-002333.4 | 0.261 | h3f3c | -0.113 |
| COX7A2 | 0.260 | cadm3 | -0.113 |
| s100a1 | 0.254 | fabp7a | -0.108 |
| alcamb | 0.252 | rnasekb | -0.104 |
| gnl3 | 0.249 | rps17 | -0.103 |
| sfrp1a | 0.247 | gapdhs | -0.102 |
| pprc1 | 0.244 | pvalb2 | -0.099 |
| npm1a | 0.243 | foxg1b | -0.098 |
| nop2 | 0.242 | pvalb1 | -0.096 |
| bms1 | 0.241 | COX3 | -0.094 |
| fbl | 0.237 | marcksl1a | -0.094 |
| dkc1 | 0.236 | actc1b | -0.092 |
| foxd5 | 0.234 | epb41a | -0.092 |
| gar1 | 0.232 | mt-atp6 | -0.091 |
| otx1 | 0.232 | atp5mc1 | -0.090 |
| nop58 | 0.232 | atp6v0cb | -0.090 |
| apoc1 | 0.228 | hbbe1.3 | -0.089 |
| LOC108190024 | 0.227 | snap25b | -0.088 |
| pkdccb | 0.224 | hbae3 | -0.088 |
| ISCU | 0.224 | btg1 | -0.087 |
| mybbp1a | 0.223 | zgc:153409 | -0.085 |
| dnmt3bb.2 | 0.223 | crx | -0.084 |