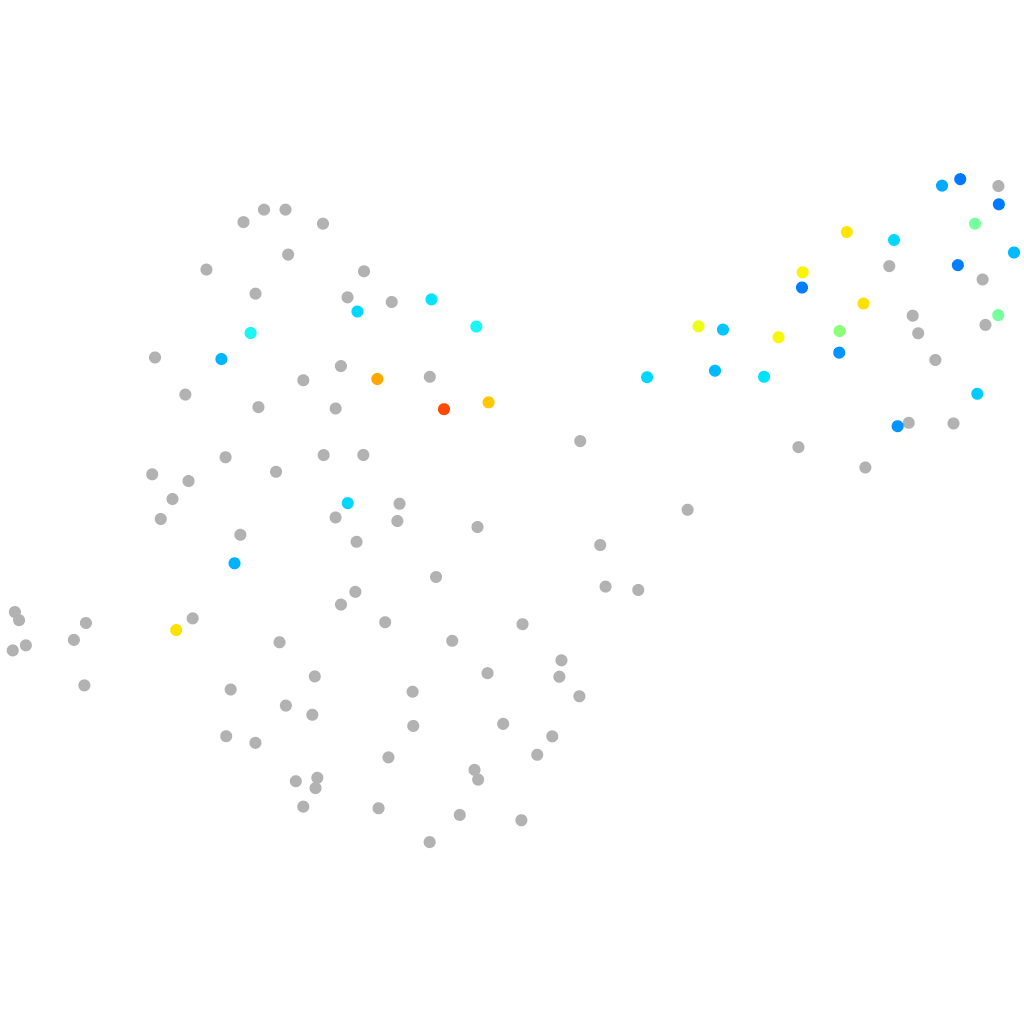
"phospholipase D family, member 6"
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| b2m | 0.689 | hnrnpaba | -0.562 |
| psme2 | 0.687 | ncl | -0.531 |
| psmb8a | 0.661 | ptmab | -0.500 |
| hyi | 0.661 | si:ch73-281n10.2 | -0.482 |
| elovl2 | 0.645 | NC-002333.4 | -0.448 |
| MCMDC2 | 0.636 | akap12b | -0.438 |
| srsf7b | 0.629 | hmgb1a | -0.433 |
| alcama | 0.620 | stm | -0.429 |
| LOC101882645 | 0.619 | marcksb | -0.422 |
| parp12a | 0.617 | hnrnpa1b | -0.420 |
| zgc:158640 | 0.617 | marcksl1b | -0.406 |
| crybg1a | 0.614 | ybx1 | -0.405 |
| zgc:63470 | 0.611 | banf1 | -0.400 |
| acaa1 | 0.609 | hnrnpa1a | -0.384 |
| si:ch211-262e15.1 | 0.608 | h1m | -0.379 |
| phb2b | 0.606 | snrpa | -0.379 |
| psme1 | 0.606 | ISCU | -0.379 |
| fam204a | 0.604 | nucks1a | -0.375 |
| hint1 | 0.602 | anp32e | -0.369 |
| CU660013.4 | 0.598 | srrt | -0.368 |
| mycbp | 0.597 | apoc1 | -0.352 |
| sdhda | 0.596 | brd3a | -0.351 |
| ldlrb | 0.592 | hmga1a | -0.345 |
| tdrkh | 0.592 | srsf1a | -0.344 |
| dusp22b | 0.589 | ppig | -0.342 |
| hsd17b7 | 0.588 | si:ch211-262h13.3 | -0.339 |
| si:ch211-194c3.5 | 0.585 | gdf3 | -0.333 |
| gchfr | 0.581 | snrpb2 | -0.332 |
| CR382383.2 | 0.581 | arf1 | -0.332 |
| calm3a | 0.581 | ccna2 | -0.332 |
| cox5b2 | 0.579 | tpx2 | -0.327 |
| sqor | 0.577 | ddit4 | -0.326 |
| actr8 | 0.574 | rbm4.3 | -0.325 |
| rab11bb | 0.573 | pttg1 | -0.325 |
| ola1 | 0.573 | nop56 | -0.324 |