"phospholipase C, eta 2b"
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line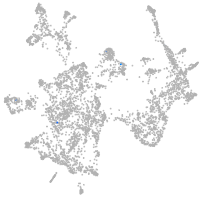 |
epidermis |
periderm |
fin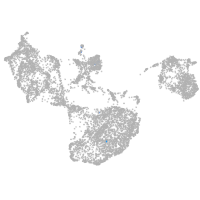 |
ionocytes / mucous |
pigment cells |
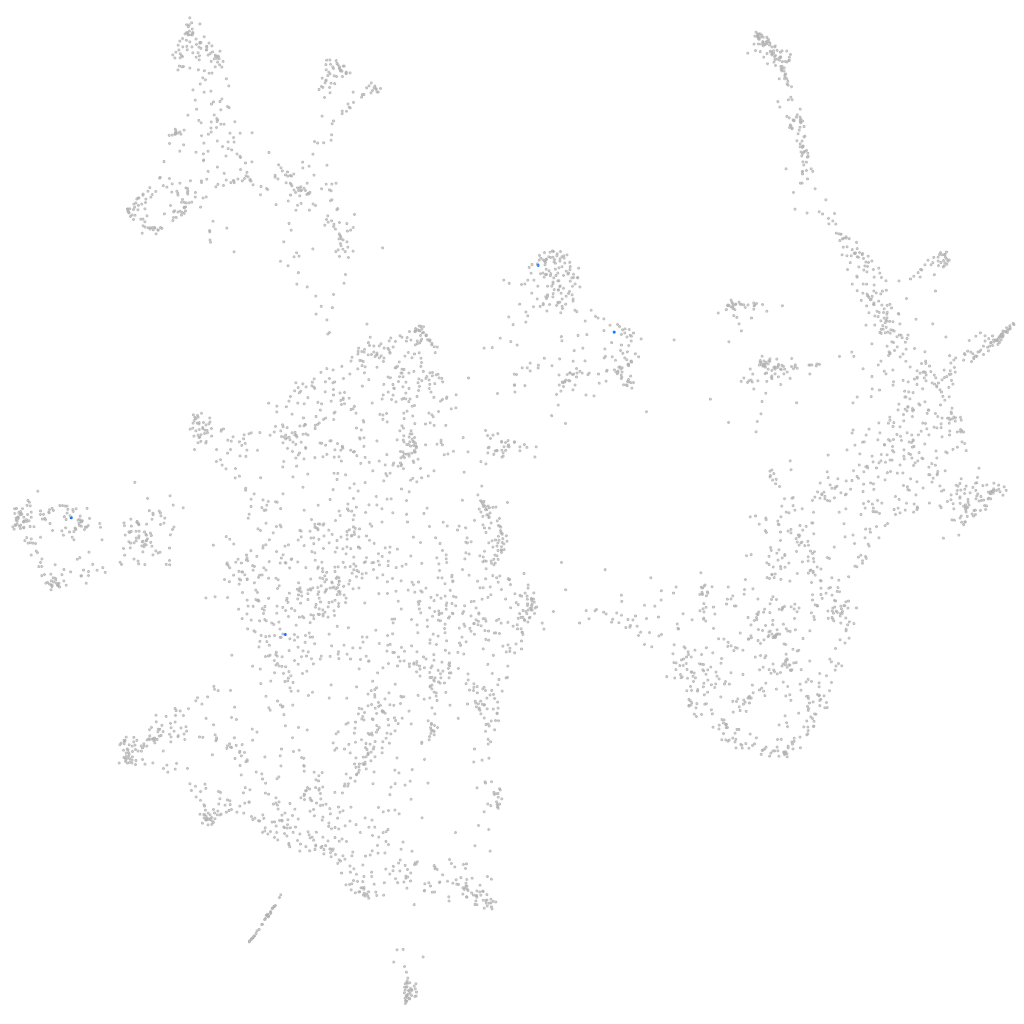
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| gys2 | 0.612 | spint2 | -0.027 |
| blk | 0.548 | mcl1b | -0.025 |
| c1qtnf5 | 0.384 | npc2 | -0.024 |
| LO018303.1 | 0.374 | mgst3b | -0.023 |
| pcdh1gb9 | 0.359 | mtpn | -0.022 |
| piwil2 | 0.331 | etfa | -0.022 |
| ankrd33aa | 0.315 | slc38a5b | -0.022 |
| rnf151 | 0.314 | ccdc124 | -0.021 |
| zmp:0000000755 | 0.309 | GCA | -0.021 |
| foxe1 | 0.308 | spint1a | -0.021 |
| XLOC-043765 | 0.307 | prrg2 | -0.021 |
| rasgrf2b | 0.303 | rnd3a | -0.020 |
| heyl | 0.292 | mrpl11 | -0.020 |
| si:ch211-106h4.12 | 0.289 | msi2b | -0.020 |
| si:dkey-283b1.7 | 0.288 | psph | -0.020 |
| enpp7.1 | 0.280 | rab6a | -0.020 |
| fam20a | 0.275 | aktip | -0.020 |
| rp1l1b | 0.267 | commd7 | -0.020 |
| tat | 0.257 | crebzf | -0.020 |
| hhex | 0.252 | serpine2 | -0.020 |
| npy | 0.251 | chd7 | -0.020 |
| si:ch211-226h8.4 | 0.248 | yy1a | -0.020 |
| si:dkey-22h13.3 | 0.229 | eya4 | -0.019 |
| camkva | 0.227 | tmem18 | -0.019 |
| LOC101885083 | 0.224 | socs3b | -0.019 |
| lrrc3b | 0.223 | si:ch211-15b10.6 | -0.019 |
| nr5a2 | 0.223 | chmp4bb | -0.019 |
| lratb.1 | 0.217 | foxp4 | -0.019 |
| si:rp71-68n21.9 | 0.214 | tmem54a | -0.019 |
| LOC103908899 | 0.206 | si:ch211-286b5.5 | -0.019 |
| dcstamp | 0.203 | litaf | -0.019 |
| si:ch211-153l6.6 | 0.203 | napab | -0.019 |
| cpne8 | 0.202 | trappc3 | -0.019 |
| loxa | 0.201 | rap1b | -0.019 |
| bglapl | 0.197 | XLOC-035413 | -0.019 |