pleiomorphic adenoma gene-like 2
ZFIN 
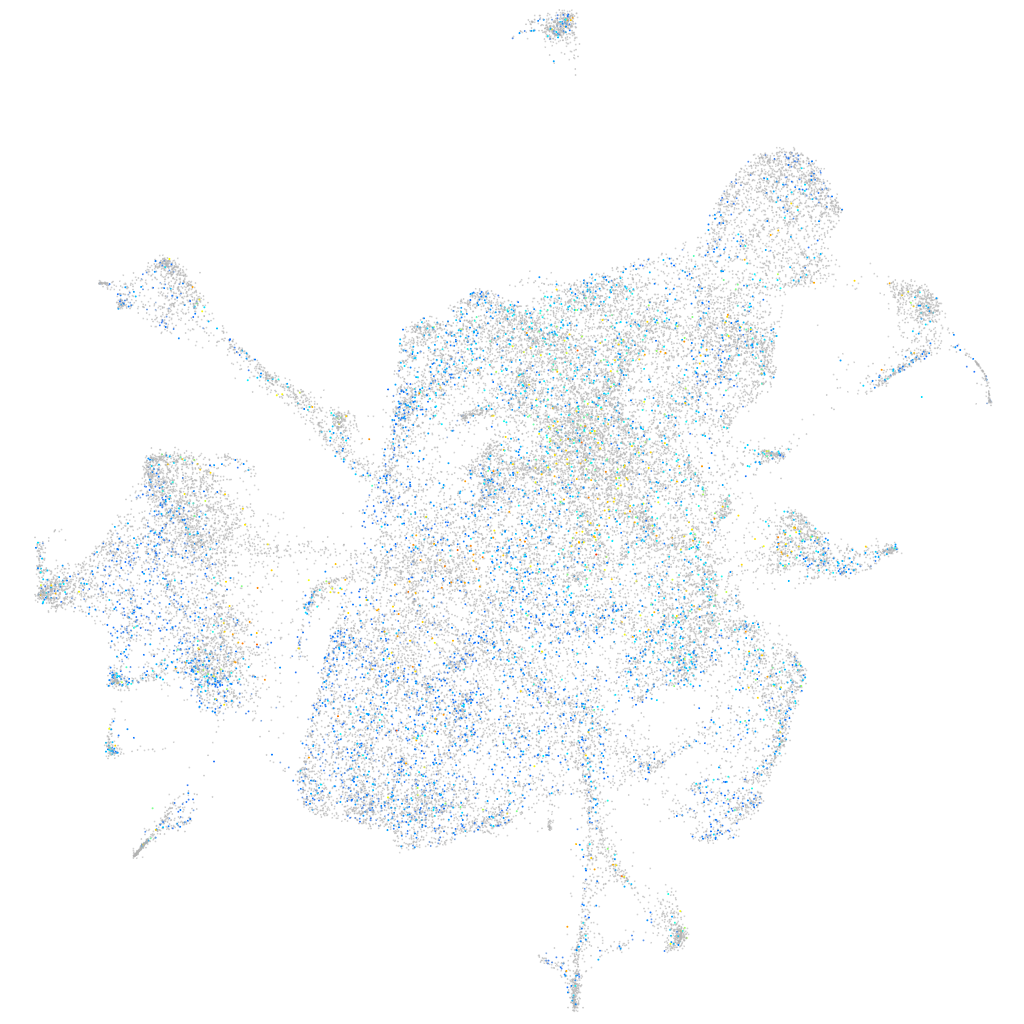
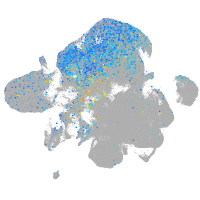
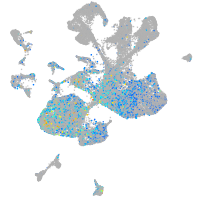
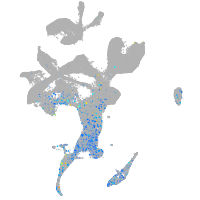
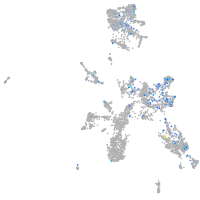
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| NC-002333.4 | 0.051 | matn1 | -0.037 |
| col5a1 | 0.050 | mia | -0.036 |
| hmga2 | 0.049 | col11a2 | -0.033 |
| col1a1b | 0.048 | wisp3 | -0.033 |
| ppiaa | 0.047 | col9a1a | -0.033 |
| fstl1b | 0.045 | ppdpfb | -0.032 |
| col1a1a | 0.045 | ucmab | -0.032 |
| mt-co1 | 0.044 | tnxba | -0.031 |
| cd81a | 0.042 | snorc | -0.031 |
| tcf12 | 0.041 | col9a3 | -0.030 |
| col5a2a | 0.040 | fgfbp2b | -0.030 |
| mdka | 0.039 | col2a1a | -0.030 |
| cilp | 0.039 | col9a2 | -0.030 |
| pleca | 0.039 | acana | -0.030 |
| boc | 0.039 | otos | -0.029 |
| CR931782.2 | 0.039 | slc25a5 | -0.029 |
| col1a2 | 0.038 | pcolceb | -0.029 |
| COX3 | 0.038 | si:dkey-19b23.8 | -0.028 |
| zgc:165555.3 | 0.038 | apoa1b | -0.028 |
| mrc2 | 0.038 | epyc | -0.027 |
| hist1h4l | 0.037 | ptgdsa | -0.026 |
| ctsk | 0.037 | VIT | -0.026 |
| zgc:158463 | 0.036 | meltf | -0.026 |
| stom | 0.036 | pdcd6 | -0.026 |
| lig1 | 0.036 | fxyd1 | -0.026 |
| postnb | 0.036 | cthrc1b | -0.025 |
| cirbpa | 0.036 | atp6v1e1b | -0.025 |
| pcdh18b | 0.035 | romo1 | -0.025 |
| rpl3 | 0.035 | rock2b | -0.025 |
| CU929237.1 | 0.035 | mcl1a | -0.025 |
| fgfr2 | 0.035 | CYTL1 | -0.025 |
| ptprsa | 0.035 | itih1 | -0.025 |
| tenm3 | 0.034 | apoa2 | -0.024 |
| marcksl1a | 0.034 | atp6v1g1 | -0.024 |
| foxp4 | 0.034 | rpl19 | -0.024 |