"phospholipase A2, group IVAb (cytosolic, calcium-dependent)"
ZFIN 
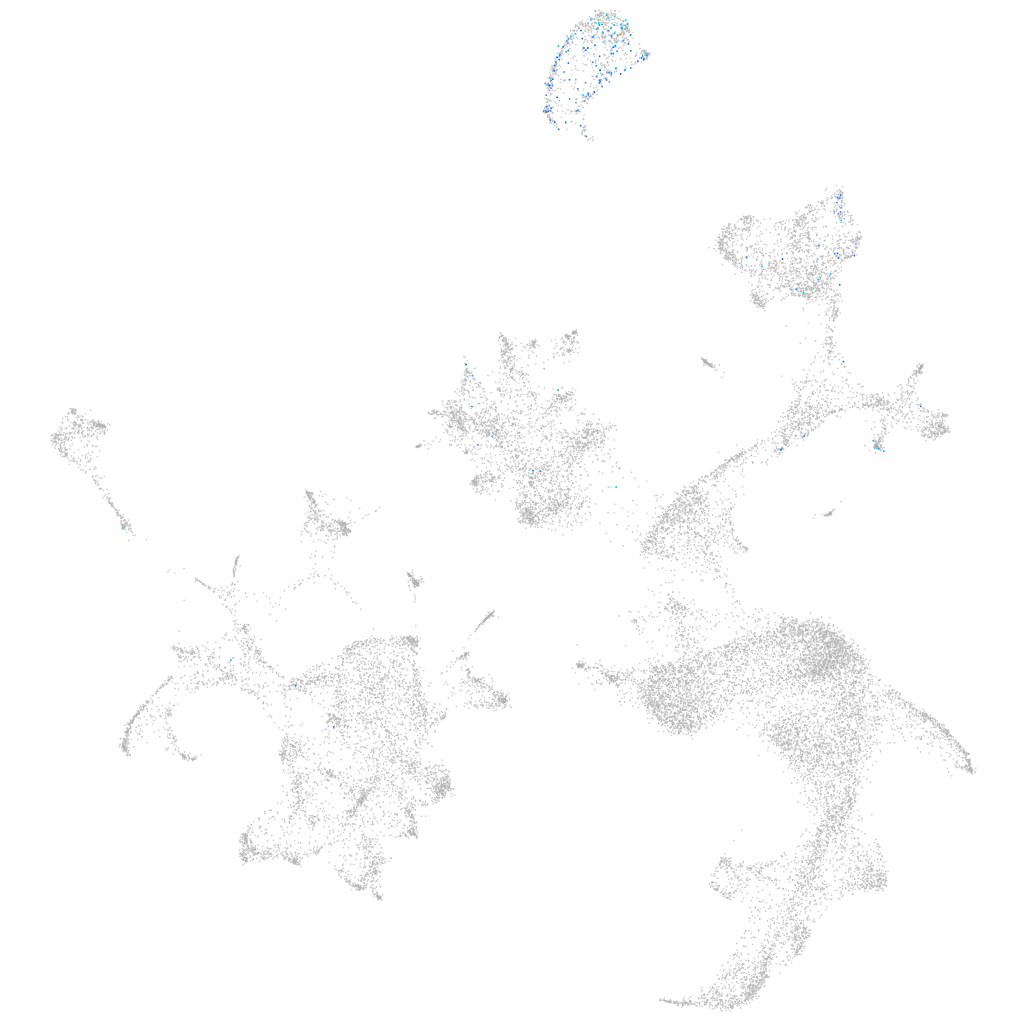
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle 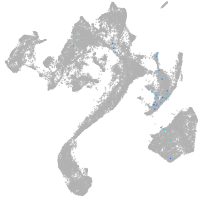 |
mural cells / non-skeletal muscle  |
mesenchyme 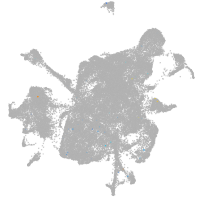 |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line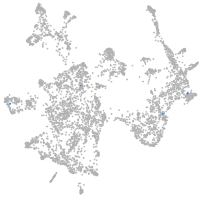 |
epidermis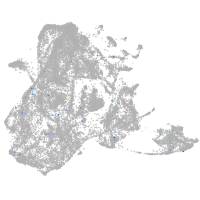 |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| ch25hl2 | 0.306 | hbae1.1 | -0.084 |
| npsn | 0.305 | hbae3 | -0.082 |
| mpx | 0.300 | hbae1.3 | -0.081 |
| lyz | 0.295 | hbbe1.3 | -0.080 |
| il34 | 0.292 | hbbe2 | -0.079 |
| ponzr6 | 0.286 | hbbe1.1 | -0.078 |
| ctss1 | 0.285 | cahz | -0.072 |
| cfbl | 0.281 | fth1a | -0.070 |
| cpa5 | 0.281 | blvrb | -0.069 |
| mmp13a | 0.280 | hbbe1.2 | -0.069 |
| scinlb | 0.275 | hemgn | -0.069 |
| si:ch1073-429i10.1 | 0.274 | lmo2 | -0.065 |
| lect2l | 0.274 | alas2 | -0.065 |
| scpp8 | 0.273 | si:ch211-250g4.3 | -0.064 |
| sms | 0.273 | slc4a1a | -0.061 |
| si:dkey-102g19.3 | 0.271 | zgc:56095 | -0.061 |
| glipr1a | 0.269 | nt5c2l1 | -0.059 |
| alox5ap | 0.266 | creg1 | -0.059 |
| prkcda | 0.262 | prdx2 | -0.058 |
| dhrs9 | 0.258 | epb41b | -0.057 |
| il6r | 0.256 | zgc:163057 | -0.056 |
| lta4h | 0.256 | si:ch211-207c6.2 | -0.055 |
| aldh8a1 | 0.254 | nmt1b | -0.053 |
| si:ch211-286b5.8 | 0.249 | aqp1a.1 | -0.051 |
| LOC101884267 | 0.249 | tmod4 | -0.051 |
| si:ch1073-443f11.2 | 0.248 | plac8l1 | -0.049 |
| vamp8 | 0.246 | selenoj | -0.047 |
| si:ch211-8c17.2 | 0.246 | sptb | -0.047 |
| gapdh | 0.246 | si:ch211-227m13.1 | -0.046 |
| ppdpfa | 0.245 | uros | -0.046 |
| si:ch211-276a23.5 | 0.242 | arl4aa | -0.045 |
| cd7al | 0.240 | tfr1a | -0.045 |
| si:dkey-238m4.4 | 0.237 | hbae5 | -0.045 |
| mmp9 | 0.233 | hdr | -0.044 |
| nccrp1 | 0.232 | rfesd | -0.044 |