"phospholipase A2, group XIIB"
ZFIN 
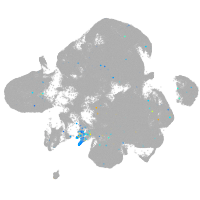
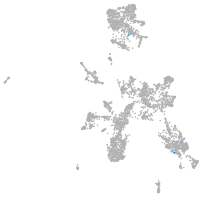
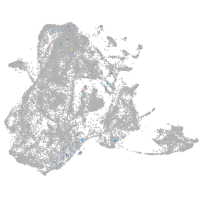
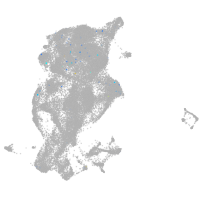
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| apobb.1 | 0.774 | epcam | -0.406 |
| afp4 | 0.768 | slc38a5b | -0.378 |
| apoc2 | 0.764 | tuba8l4 | -0.361 |
| apoa4b.1 | 0.750 | si:ch211-222l21.1 | -0.353 |
| gpx4a | 0.736 | cirbpa | -0.350 |
| fbp1b | 0.656 | ywhaz | -0.345 |
| slc27a2a | 0.638 | cldnb | -0.343 |
| cdo1 | 0.630 | ptmab | -0.333 |
| gapdh | 0.629 | hmgb2a | -0.331 |
| suclg1 | 0.626 | ier2b | -0.331 |
| apoa1a | 0.622 | h2afvb | -0.329 |
| rbp2a | 0.610 | si:ch73-1a9.3 | -0.322 |
| gamt | 0.604 | cldn7b | -0.321 |
| apoa1b | 0.601 | hmgb3a | -0.320 |
| rdh1 | 0.600 | idh2 | -0.319 |
| dgat2 | 0.600 | khdrbs1a | -0.317 |
| fabp2 | 0.597 | CR383676.1 | -0.317 |
| dhrs9 | 0.595 | her9 | -0.310 |
| aldob | 0.594 | syncrip | -0.306 |
| mttp | 0.592 | cirbpb | -0.304 |
| scp2a | 0.592 | hmgn2 | -0.304 |
| glud1b | 0.580 | hmgb2b | -0.303 |
| tpi1b | 0.575 | nucks1a | -0.303 |
| fabp1b.1 | 0.561 | hnrnpaba | -0.303 |
| suclg2 | 0.560 | h3f3d | -0.302 |
| mdh1aa | 0.559 | hmgn6 | -0.300 |
| srd5a2a | 0.557 | jpt1b | -0.300 |
| sult2st2 | 0.556 | si:ch211-195b11.3 | -0.297 |
| gcshb | 0.555 | pnrc2 | -0.297 |
| faxdc2 | 0.555 | icn | -0.297 |
| apoa4b.2 | 0.553 | bcam | -0.293 |
| ugt1a7 | 0.553 | her6 | -0.291 |
| slc37a4a | 0.551 | agr2 | -0.290 |
| mgst1.2 | 0.549 | stard10 | -0.290 |
| agmo | 0.547 | npdc1a | -0.289 |