piwi-like RNA-mediated gene silencing 2
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs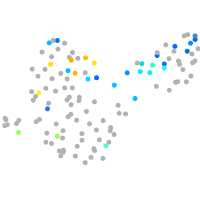 |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| piwil1 | 0.696 | hnrnpaba | -0.516 |
| psmb8a | 0.670 | hmga1a | -0.444 |
| LOC100149647 | 0.659 | anp32e | -0.415 |
| smc1b | 0.629 | ybx1 | -0.412 |
| cox5b2 | 0.623 | actb1 | -0.385 |
| elavl4 | 0.608 | cx43.4 | -0.368 |
| tuba7l | 0.606 | h1m | -0.363 |
| cks2 | 0.604 | ccna2 | -0.360 |
| hormad1 | 0.583 | rbm7 | -0.355 |
| stag3 | 0.582 | snrpa | -0.351 |
| LOC101885930 | 0.573 | si:ch73-281n10.2 | -0.345 |
| atr | 0.558 | akap12b | -0.343 |
| CR382383.2 | 0.557 | hmgb2a | -0.340 |
| gchfr | 0.553 | ptmab | -0.331 |
| adad1 | 0.552 | hnrnpa1b | -0.327 |
| tdrd9 | 0.549 | ncl | -0.325 |
| tdrd1 | 0.547 | hnrnpub | -0.323 |
| kcnv2b | 0.542 | myl12.1 | -0.323 |
| carm1l | 0.542 | hmgb1a | -0.318 |
| LOC101883356 | 0.542 | id1 | -0.316 |
| CU660013.4 | 0.539 | stm | -0.313 |
| psme2 | 0.539 | kif23 | -0.308 |
| actc1c | 0.539 | nucks1a | -0.305 |
| psme1 | 0.539 | syncrip | -0.301 |
| zgc:92591 | 0.537 | cd2bp2 | -0.301 |
| CU929391.1 | 0.535 | hspb1 | -0.301 |
| oxr1a | 0.532 | nucks1b | -0.300 |
| fam102ba | 0.529 | ctsla | -0.294 |
| LOC101885829 | 0.525 | ddit4 | -0.291 |
| LOC101886261 | 0.525 | safb | -0.287 |
| cpne2 | 0.523 | apoc1 | -0.285 |
| alcama | 0.522 | zgc:110216 | -0.282 |
| fkbp6 | 0.519 | zgc:110425 | -0.282 |
| tdrkh | 0.519 | histh1l | -0.280 |
| ost4 | 0.517 | sdc4 | -0.280 |