"phosphoinositide-3-kinase, regulatory subunit 3b (gamma)"
ZFIN 
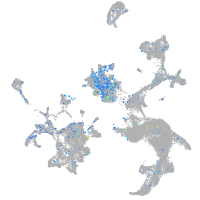
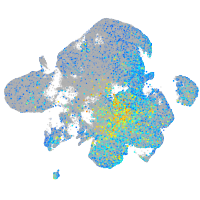
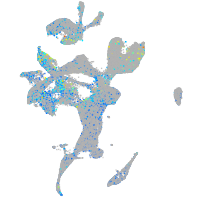
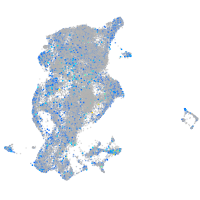
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| phactr3a | 0.285 | slc25a5 | -0.174 |
| mab21l2 | 0.282 | atp1b1a | -0.149 |
| XLOC-035847 | 0.254 | atp5f1c | -0.141 |
| meis2a | 0.253 | rps2 | -0.141 |
| stmn1b | 0.253 | rpl35a | -0.139 |
| CACNA2D1 | 0.252 | atp5pd | -0.137 |
| si:ch73-386h18.1 | 0.246 | atp5mc3b | -0.134 |
| slit1b | 0.243 | cdh17 | -0.131 |
| st8sia5 | 0.238 | cyc1 | -0.130 |
| CABZ01078320.1 | 0.232 | COX5B | -0.130 |
| robo2 | 0.230 | ndrg1a | -0.130 |
| nova2 | 0.229 | cox6a1 | -0.130 |
| tmem179 | 0.226 | atp5pf | -0.128 |
| pbx3b | 0.226 | cox4i1 | -0.127 |
| clvs2 | 0.220 | ndufb8 | -0.124 |
| elavl4 | 0.220 | atp5f1d | -0.123 |
| kif3cb | 0.219 | suclg1 | -0.123 |
| apc2 | 0.218 | chp1 | -0.122 |
| marcksl1b | 0.216 | krt8 | -0.118 |
| fam171b | 0.213 | mdh2 | -0.118 |
| elavl3 | 0.212 | krt18a.1 | -0.118 |
| gpr85 | 0.212 | atp5l | -0.117 |
| gpm6aa | 0.211 | slc25a3b | -0.117 |
| sncb | 0.211 | cox5aa | -0.116 |
| csdc2a | 0.210 | rpl37 | -0.116 |
| tshz2 | 0.209 | atp5fa1 | -0.116 |
| gap43 | 0.209 | atp1a1a.4 | -0.115 |
| gpm6ab | 0.206 | rps10 | -0.115 |
| LOC110438594 | 0.205 | uqcrfs1 | -0.115 |
| tmem178b | 0.203 | vdac2 | -0.115 |
| si:ch211-150g13.3 | 0.203 | uqcrq | -0.115 |
| map4l | 0.202 | cox8a | -0.114 |
| jpt1a | 0.202 | prkab1b | -0.114 |
| sema4gb | 0.201 | mycbp | -0.114 |
| nmnat2 | 0.200 | mpc1 | -0.114 |