phosphoinositide-3-kinase interacting protein 1
ZFIN 
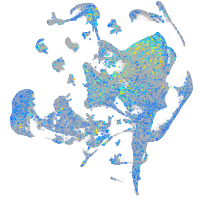
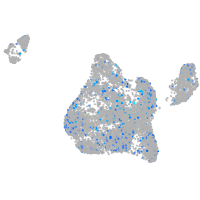
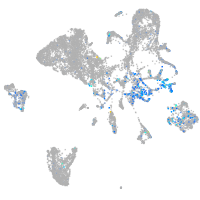
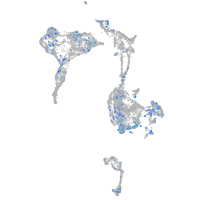
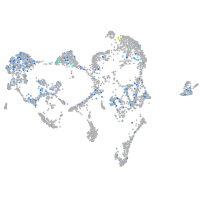
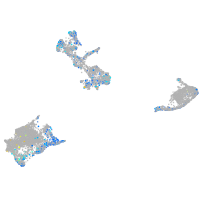
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| ndufa4l2a | 0.292 | gapdh | -0.227 |
| scgn | 0.280 | ahcy | -0.214 |
| pax6b | 0.278 | gamt | -0.194 |
| SLC5A10 | 0.277 | eno3 | -0.190 |
| isl1 | 0.274 | gatm | -0.184 |
| kcnj11 | 0.270 | mat1a | -0.176 |
| ins | 0.260 | aldob | -0.174 |
| scg3 | 0.257 | glud1b | -0.172 |
| si:dkey-153k10.9 | 0.255 | gstt1a | -0.172 |
| neurod1 | 0.253 | fbp1b | -0.172 |
| ptn | 0.250 | cx32.3 | -0.169 |
| rprmb | 0.249 | apoa4b.1 | -0.165 |
| pcsk1nl | 0.248 | bhmt | -0.163 |
| cntnap2a | 0.242 | suclg1 | -0.163 |
| pcsk2 | 0.241 | apoc2 | -0.162 |
| slc30a2 | 0.239 | nupr1b | -0.161 |
| tspan7b | 0.236 | scp2a | -0.159 |
| insm1a | 0.231 | sod1 | -0.157 |
| vat1 | 0.227 | apoa1b | -0.157 |
| pcsk1 | 0.226 | gpx4a | -0.155 |
| rims2a | 0.224 | sod2 | -0.152 |
| lysmd2 | 0.221 | sult2st2 | -0.151 |
| gcgb | 0.220 | gcshb | -0.149 |
| tspan7 | 0.220 | gnmt | -0.149 |
| gdf6a | 0.218 | afp4 | -0.148 |
| CABZ01075068.1 | 0.215 | agxtb | -0.147 |
| abhd15a | 0.215 | aldh6a1 | -0.147 |
| dmtn | 0.214 | agxta | -0.147 |
| mir7a-1 | 0.213 | rdh1 | -0.147 |
| camk1da | 0.213 | chchd10 | -0.145 |
| jpt1b | 0.212 | nipsnap3a | -0.144 |
| fam49bb | 0.211 | aldh7a1 | -0.144 |
| fstl1a | 0.210 | cox6b1 | -0.144 |
| si:dkey-42i9.4 | 0.209 | sdr16c5b | -0.143 |
| zgc:92818 | 0.209 | suclg2 | -0.143 |