PIH1 domain containing 2
ZFIN 
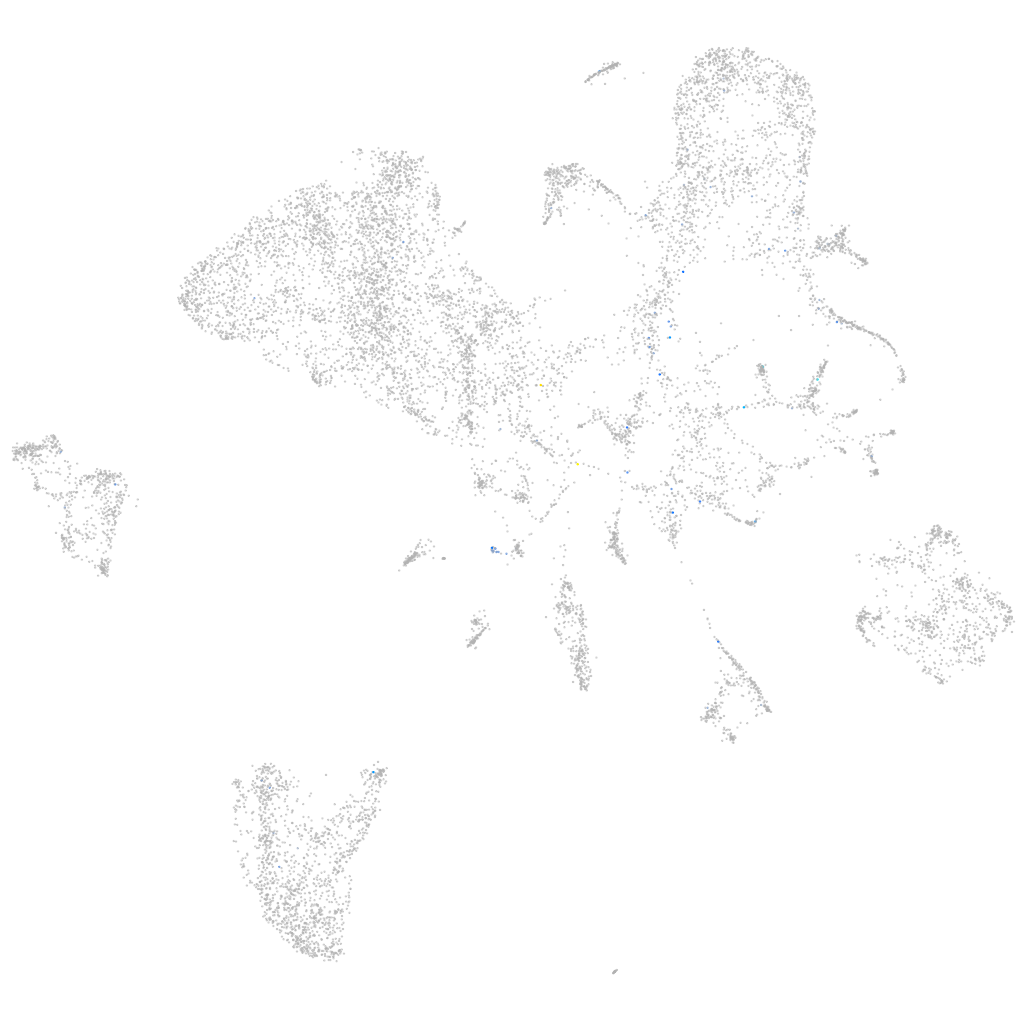
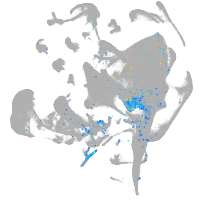
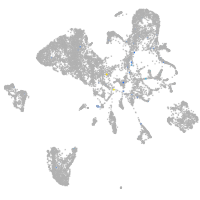
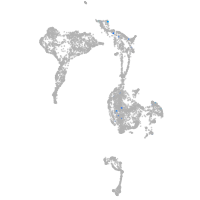
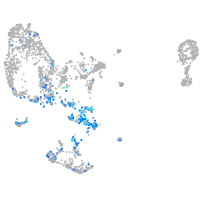
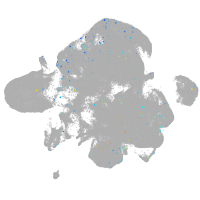
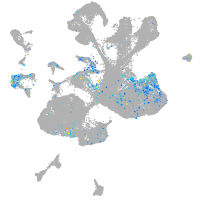
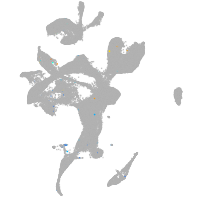
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| ldlrad4b | 0.341 | gamt | -0.046 |
| cfap126 | 0.297 | gapdh | -0.045 |
| chrna10a | 0.277 | gatm | -0.043 |
| crocc2 | 0.254 | apoc1 | -0.042 |
| ccdc83 | 0.238 | bhmt | -0.038 |
| si:dkey-108k21.27 | 0.233 | pnp4b | -0.037 |
| CU915770.1 | 0.232 | tfa | -0.037 |
| cfap52 | 0.228 | fabp3 | -0.037 |
| CR392012.2 | 0.213 | afp4 | -0.037 |
| alkal2b | 0.210 | apoa1b | -0.037 |
| si:dkey-124l13.1 | 0.209 | fabp10a | -0.036 |
| opn6b | 0.190 | apom | -0.036 |
| ccl38.1 | 0.184 | rbp2b | -0.036 |
| zgc:193811 | 0.181 | apoa4b.1 | -0.036 |
| tbata | 0.173 | serpina1 | -0.036 |
| ak9 | 0.168 | serpina1l | -0.036 |
| ccdc151 | 0.167 | scp2a | -0.036 |
| fgf18a | 0.165 | agxtb | -0.036 |
| peli1a | 0.164 | fetub | -0.035 |
| si:dkey-27p23.3 | 0.164 | grhprb | -0.035 |
| tekt4 | 0.162 | ces2 | -0.035 |
| LOC103912003 | 0.162 | mgst1.2 | -0.035 |
| KCNK12 | 0.155 | fgb | -0.035 |
| si:dkey-29j8.2 | 0.150 | apobb.1 | -0.034 |
| nsmfb | 0.145 | zgc:123103 | -0.034 |
| CR385078.1 | 0.145 | gpx4a | -0.034 |
| tekt2 | 0.144 | fgg | -0.034 |
| CABZ01076996.1 | 0.143 | c9 | -0.034 |
| rfx4 | 0.142 | apoa2 | -0.034 |
| drc1 | 0.142 | fga | -0.034 |
| cfap45 | 0.140 | LOC110437731 | -0.033 |
| si:ch211-248e11.2 | 0.139 | hpda | -0.033 |
| meig1 | 0.138 | ambp | -0.033 |
| FQ311924.1 | 0.135 | igfbp2a | -0.033 |
| FP236331.1 | 0.133 | zgc:112265 | -0.033 |