paired like homeobox 2A
ZFIN 
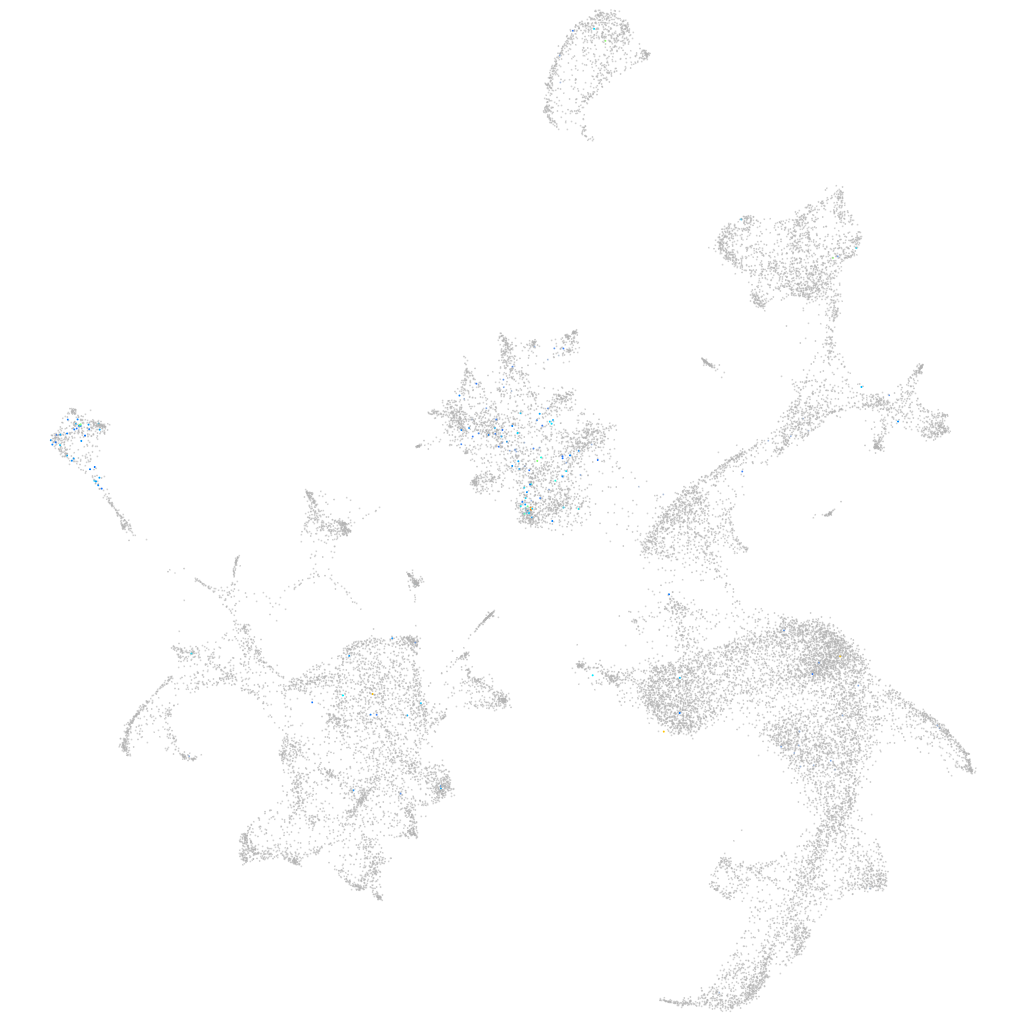
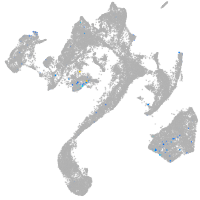
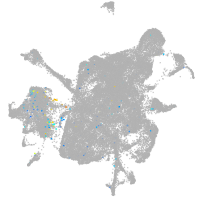
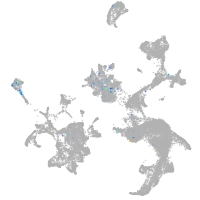
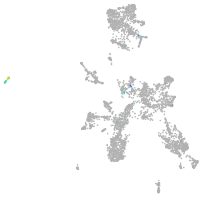
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| phox2bb | 0.385 | lmo2 | -0.036 |
| CR812832.1 | 0.318 | rps17 | -0.028 |
| prph | 0.271 | rps10 | -0.027 |
| scn1laa | 0.269 | prdx2 | -0.027 |
| tppp2 | 0.210 | aqp1a.1 | -0.027 |
| ret | 0.177 | fam210b | -0.026 |
| CR354372.1 | 0.167 | ap2m1a | -0.026 |
| isl2a | 0.166 | vdac2 | -0.025 |
| tlx2 | 0.163 | mcl1a | -0.025 |
| p2rx2 | 0.146 | hbbe3 | -0.025 |
| vip | 0.144 | tmem14ca | -0.025 |
| si:dkeyp-51f12.2 | 0.144 | slc11a2 | -0.025 |
| chrna5 | 0.138 | gata1a | -0.024 |
| ank2a | 0.137 | blvrb | -0.024 |
| htr3b | 0.136 | rpl37 | -0.024 |
| pcbp4 | 0.132 | znfl2a | -0.024 |
| epha4b | 0.132 | snx5 | -0.023 |
| st8sia2 | 0.131 | hmbsa | -0.023 |
| slc18a3a | 0.126 | prelid3b | -0.023 |
| si:ch211-272b8.6 | 0.125 | arl4aa | -0.022 |
| onecut1 | 0.124 | romo1 | -0.022 |
| elavl3 | 0.122 | TCIM (1 of many) | -0.022 |
| CABZ01003344.1 | 0.119 | tspo | -0.022 |
| si:ch211-181l5.2 | 0.118 | urod | -0.021 |
| stmn2a | 0.118 | clic2 | -0.021 |
| dlx2b | 0.117 | mob1a | -0.021 |
| elavl4 | 0.116 | egfl7 | -0.021 |
| capn1b | 0.116 | zgc:163080 | -0.021 |
| htr3a | 0.115 | nmt1b | -0.021 |
| LO018324.1 | 0.115 | cpox | -0.020 |
| kif26bb | 0.113 | uros | -0.020 |
| kif1aa | 0.112 | clec14a | -0.020 |
| fam155b | 0.112 | tal1 | -0.020 |
| si:ch1073-44g3.1 | 0.111 | cldng | -0.020 |
| gap43 | 0.111 | rhocb | -0.020 |