prohibitin 2a
ZFIN 
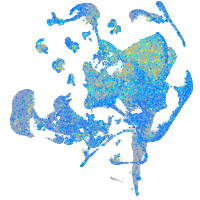
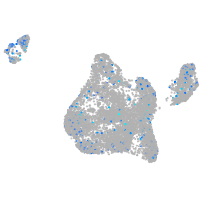
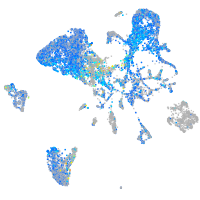
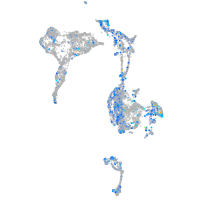
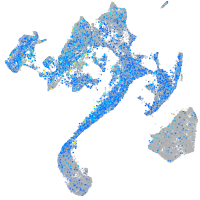
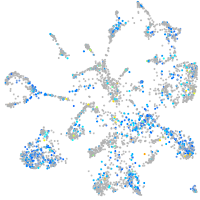
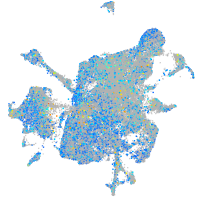
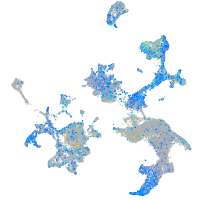
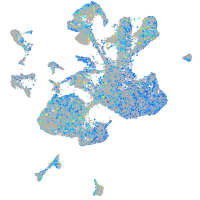
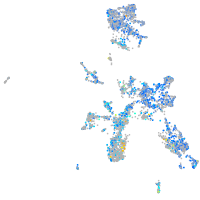
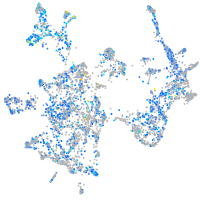
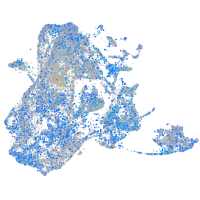
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| suclg1 | 0.321 | hspb1 | -0.278 |
| mt-atp6 | 0.317 | wu:fb97g03 | -0.268 |
| atp5f1b | 0.311 | apoeb | -0.252 |
| mt-nd1 | 0.310 | apoc1 | -0.245 |
| ldhba | 0.309 | hmgn6 | -0.234 |
| eno3 | 0.309 | hmgb2a | -0.229 |
| si:ch211-139a5.9 | 0.307 | hmga1a | -0.228 |
| sod1 | 0.304 | acin1a | -0.219 |
| elovl1b | 0.303 | nucks1a | -0.217 |
| atp6v1e1b | 0.301 | cx43.4 | -0.217 |
| mt-nd2 | 0.301 | cdx4 | -0.216 |
| cdh17 | 0.299 | si:ch73-281n10.2 | -0.214 |
| hadh | 0.298 | si:ch73-1a9.3 | -0.211 |
| atp5mc1 | 0.294 | zgc:110425 | -0.209 |
| cox5aa | 0.293 | akap12b | -0.208 |
| glud1b | 0.292 | marcksb | -0.206 |
| gstp1 | 0.291 | znfl2a | -0.205 |
| ppifb | 0.289 | anp32a | -0.203 |
| atp5pd | 0.288 | ilf3b | -0.198 |
| mt-nd4 | 0.287 | smc1al | -0.195 |
| chchd10 | 0.286 | tardbp | -0.195 |
| atp5fa1 | 0.286 | hmgb2b | -0.192 |
| gcshb | 0.284 | hnrnpaba | -0.190 |
| cox7a2a | 0.284 | seta | -0.188 |
| atp5l | 0.282 | marcksl1b | -0.188 |
| ctsla | 0.282 | zmp:0000000624 | -0.187 |
| npl | 0.281 | hnrnpabb | -0.187 |
| minos1 | 0.281 | lbx2 | -0.187 |
| atp5f1d | 0.280 | fbl | -0.187 |
| suclg2 | 0.280 | ppig | -0.186 |
| COX3 | 0.280 | BX927258.1 | -0.186 |
| aldh7a1 | 0.280 | lmo2 | -0.185 |
| prdx3 | 0.280 | lig1 | -0.185 |
| mdh1aa | 0.279 | cxcl12a | -0.184 |
| ndufs4 | 0.278 | id3 | -0.183 |