phosphatase and actin regulator 4a
ZFIN 
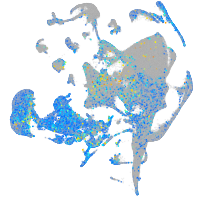
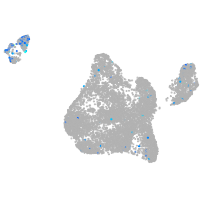
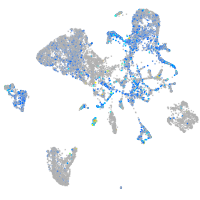
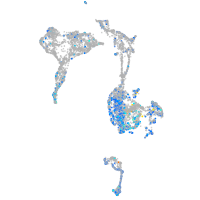
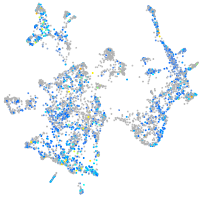
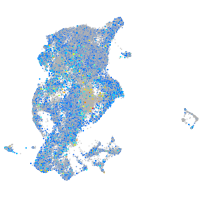
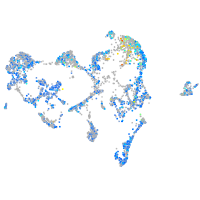
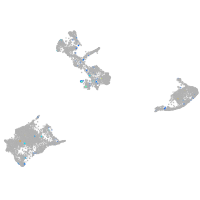
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| podxl | 0.430 | BX088707.3 | -0.212 |
| jam2b | 0.416 | tpm1 | -0.157 |
| krt94 | 0.408 | inka1a | -0.149 |
| cd151 | 0.402 | acta2 | -0.146 |
| tmem88b | 0.353 | tagln | -0.144 |
| cavin2b | 0.340 | gapdhs | -0.130 |
| si:ch211-156j16.1 | 0.333 | myh11a | -0.129 |
| gstm.3 | 0.333 | pdgfrb | -0.121 |
| cav1 | 0.324 | mylkb | -0.120 |
| krt8 | 0.315 | lamc3 | -0.119 |
| aqp1a.1 | 0.310 | lmod1b | -0.118 |
| cavin1b | 0.303 | myl9a | -0.118 |
| krt15 | 0.296 | ak1 | -0.118 |
| ftr82 | 0.294 | pcdh18a | -0.117 |
| bcam | 0.290 | lbh | -0.113 |
| bnc2 | 0.283 | mylka | -0.111 |
| rhag | 0.264 | rasl12 | -0.111 |
| tmem98 | 0.263 | barx1 | -0.107 |
| hspb1 | 0.263 | foxl1 | -0.105 |
| tgm2b | 0.261 | ndufa4l2a | -0.104 |
| scarb2a | 0.258 | ecrg4a | -0.104 |
| tmem88a | 0.257 | cox4i2 | -0.103 |
| myrf | 0.255 | notch3 | -0.102 |
| ehd1b | 0.252 | tpi1b | -0.100 |
| COLEC10 | 0.252 | s1pr5a | -0.100 |
| colec11 | 0.249 | igfbp7 | -0.100 |
| wt1b | 0.247 | angptl5 | -0.098 |
| mmel1 | 0.245 | zgc:158343 | -0.098 |
| tjp2b | 0.244 | rem1 | -0.098 |
| dag1 | 0.244 | wnt11r | -0.096 |
| gata6 | 0.240 | mgll | -0.095 |
| wt1a | 0.232 | myocd | -0.091 |
| jcada | 0.231 | calm2a | -0.091 |
| arl4aa | 0.228 | foxf2b | -0.090 |
| krt18a.1 | 0.225 | eno3 | -0.090 |