phosphogluconate dehydrogenase
ZFIN 
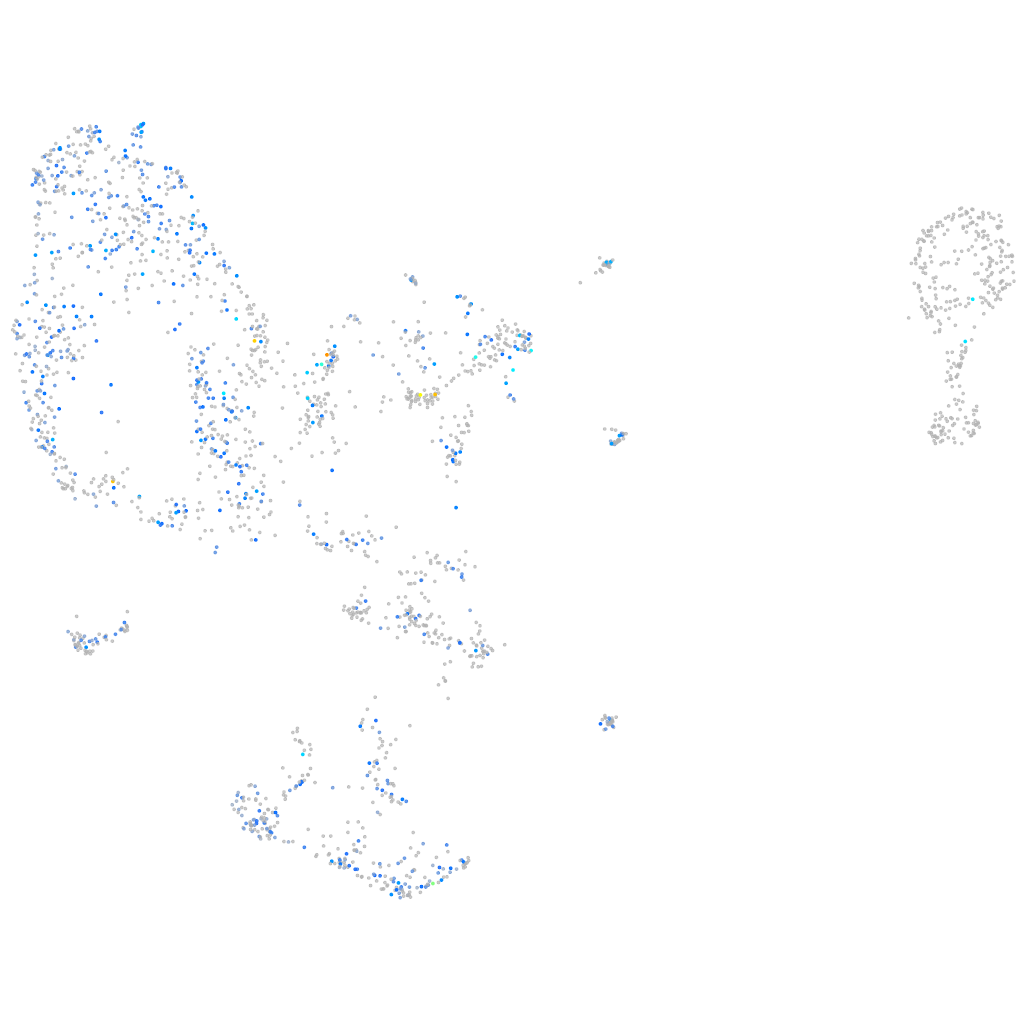
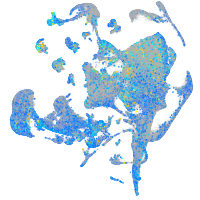
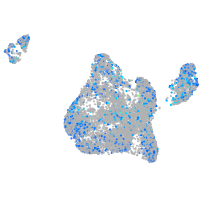
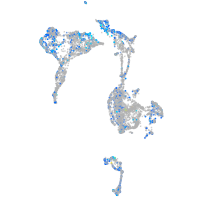
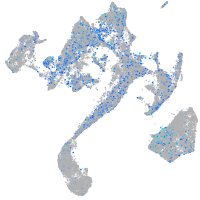
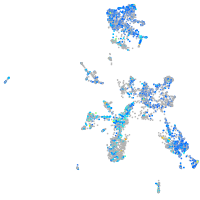
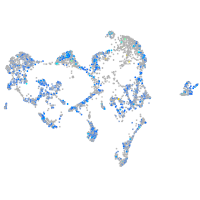
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| atp6v1e1b | 0.224 | hspb1 | -0.194 |
| suclg1 | 0.219 | wu:fb97g03 | -0.192 |
| eno3 | 0.214 | si:ch73-1a9.3 | -0.178 |
| atp5fa1 | 0.212 | apoc1 | -0.177 |
| aldh7a1 | 0.212 | hnrnpa1b | -0.176 |
| tmprss2 | 0.211 | anp32a | -0.170 |
| gcshb | 0.210 | hmgb2a | -0.163 |
| ctsla | 0.209 | ppig | -0.161 |
| aco1 | 0.207 | nucks1a | -0.161 |
| prdx2 | 0.207 | apoeb | -0.160 |
| gstp1 | 0.206 | seta | -0.158 |
| GCA | 0.205 | ilf3b | -0.158 |
| suclg2 | 0.204 | cx43.4 | -0.156 |
| etfa | 0.204 | hmga1a | -0.156 |
| ddt | 0.202 | si:ch211-222l21.1 | -0.155 |
| upb1 | 0.202 | cdx4 | -0.152 |
| scrn2 | 0.201 | znfl2a | -0.151 |
| atp5f1b | 0.201 | acin1a | -0.151 |
| tpi1b | 0.199 | hmgb2b | -0.150 |
| ezra | 0.199 | hnrnpaba | -0.150 |
| mt-atp6 | 0.198 | snrnp70 | -0.147 |
| ehd1b | 0.198 | ptmab | -0.147 |
| elovl1b | 0.198 | lig1 | -0.145 |
| eps8l3a | 0.198 | lmo2 | -0.145 |
| ldhba | 0.197 | zmp:0000000624 | -0.143 |
| gapdh | 0.196 | si:ch73-281n10.2 | -0.142 |
| cryl1 | 0.195 | dkc1 | -0.142 |
| ca2 | 0.195 | ncl | -0.142 |
| ctsd | 0.194 | akap12b | -0.142 |
| zgc:101040 | 0.194 | zgc:110425 | -0.142 |
| atp1b1a | 0.193 | gnl3 | -0.140 |
| smdt1b | 0.193 | anp32b | -0.139 |
| hadh | 0.193 | fbl | -0.139 |
| eps8l3b | 0.192 | hmgn6 | -0.139 |
| ahcyl1 | 0.192 | rbmx2 | -0.139 |