"6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1"
ZFIN 
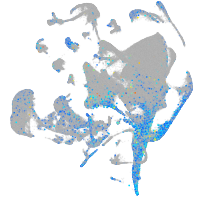
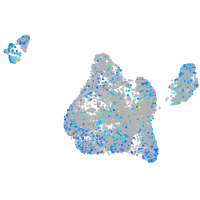
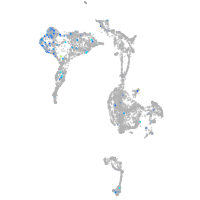
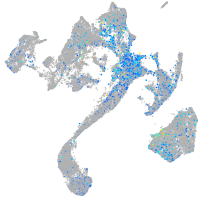
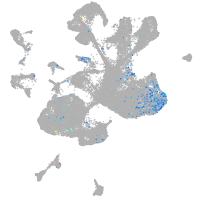
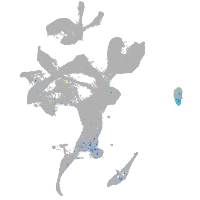
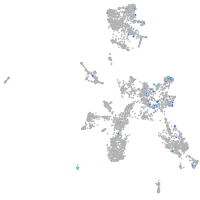
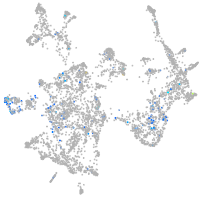
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| hspb1 | 0.332 | ptmaa | -0.150 |
| shisa2a | 0.307 | CR383676.1 | -0.148 |
| si:ch211-152c2.3 | 0.298 | rpl37 | -0.143 |
| stm | 0.295 | rps10 | -0.138 |
| cyp26a1 | 0.291 | h3f3a | -0.131 |
| apoeb | 0.283 | ppiab | -0.124 |
| aldob | 0.277 | zgc:114188 | -0.123 |
| hesx1 | 0.277 | ckbb | -0.114 |
| s100a1 | 0.265 | COX3 | -0.110 |
| LOC108190024 | 0.264 | gpm6aa | -0.109 |
| zmp:0000000624 | 0.264 | tuba1c | -0.109 |
| polr3gla | 0.262 | si:ch1073-429i10.3.1 | -0.108 |
| pprc1 | 0.259 | gpm6ab | -0.102 |
| tdgf1 | 0.258 | marcksl1a | -0.101 |
| pkdccb | 0.257 | h3f3c | -0.099 |
| sox19a | 0.253 | hnrnpa0l | -0.099 |
| asb11 | 0.249 | zgc:158463 | -0.098 |
| npm1a | 0.245 | cadm3 | -0.097 |
| COX7A2 | 0.241 | mt-atp6 | -0.097 |
| nop2 | 0.240 | rps17 | -0.094 |
| fbl | 0.238 | mt-co2 | -0.092 |
| bms1 | 0.237 | mt-nd1 | -0.088 |
| dkc1 | 0.237 | pvalb1 | -0.088 |
| nr6a1a | 0.235 | rnasekb | -0.088 |
| nop58 | 0.234 | pvalb2 | -0.085 |
| gar1 | 0.232 | fabp7a | -0.084 |
| foxd5 | 0.232 | gapdhs | -0.082 |
| gnl3 | 0.231 | atp6v0cb | -0.081 |
| rsl1d1 | 0.226 | zgc:153409 | -0.080 |
| pes | 0.224 | hbbe1.3 | -0.080 |
| apela | 0.224 | actc1b | -0.078 |
| admp | 0.222 | epb41a | -0.078 |
| ISCU | 0.220 | foxg1b | -0.078 |
| NC-002333.4 | 0.220 | atp6v1g1 | -0.076 |
| mybbp1a | 0.220 | calm1a | -0.076 |