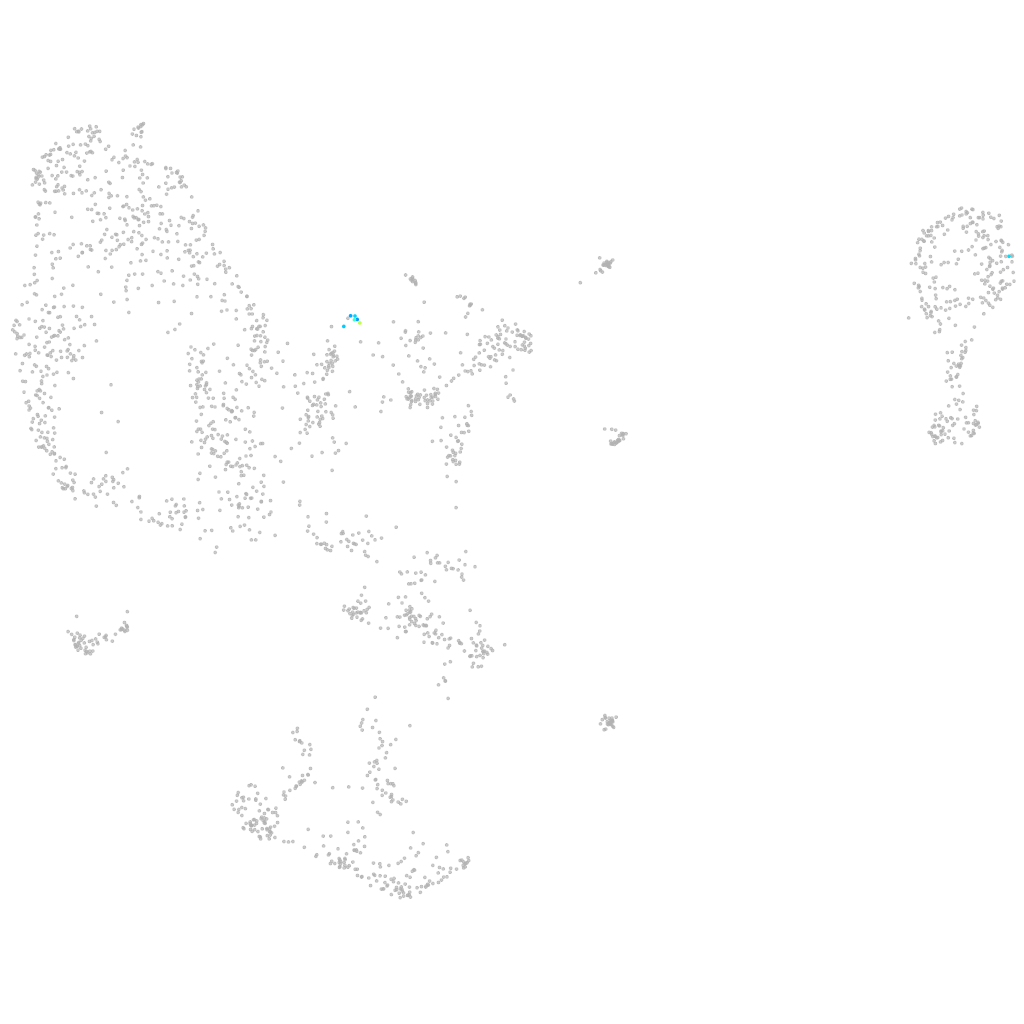
platelet/endothelial cell adhesion molecule 1
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| si:ch211-14k19.8 | 0.734 | cd9b | -0.052 |
| ptprb | 0.734 | f11r.1 | -0.044 |
| agtr1b | 0.733 | cldnc | -0.043 |
| kdr | 0.725 | abracl | -0.042 |
| tie1 | 0.707 | tomm22 | -0.041 |
| adra1d | 0.686 | cldnh | -0.040 |
| cdh5 | 0.686 | ammecr1 | -0.038 |
| calcrlb | 0.674 | cox7a2l | -0.038 |
| hspa12b | 0.660 | hnf1ba | -0.038 |
| si:dkeyp-97a10.2 | 0.656 | sf3b5 | -0.038 |
| rca2.2 | 0.636 | zgc:175088 | -0.037 |
| si:ch211-33e4.2 | 0.629 | tomm40l | -0.037 |
| myct1a | 0.624 | taf10 | -0.037 |
| myct1b | 0.623 | mycbp | -0.036 |
| ptger1b | 0.613 | prmt1 | -0.036 |
| kdrl | 0.601 | hoxb3a | -0.036 |
| si:ch73-334d15.2 | 0.594 | mrpl27 | -0.035 |
| arhgap31 | 0.588 | meig1 | -0.035 |
| arhgef9b | 0.586 | dlat | -0.035 |
| plvapb | 0.585 | ccdc47 | -0.035 |
| limch1b | 0.580 | cox5aa | -0.035 |
| cd81b | 0.576 | eif6 | -0.034 |
| lyve1b | 0.575 | rbbp4 | -0.034 |
| flt1 | 0.575 | elf3 | -0.034 |
| exoc3l2a | 0.568 | si:ch211-288g17.3 | -0.034 |
| si:ch211-145b13.6 | 0.568 | lama5 | -0.034 |
| zgc:172122 | 0.564 | emc2 | -0.034 |
| adgrl4 | 0.559 | frem2b | -0.034 |
| plvapa | 0.547 | ndrg3a | -0.034 |
| LOC101882770 | 0.543 | adi1 | -0.034 |
| myrfl | 0.541 | sstr5 | -0.034 |
| ecscr | 0.538 | stx12 | -0.034 |
| LOC108182616 | 0.537 | si:ch211-193l2.10 | -0.033 |
| gpr182 | 0.533 | snrpe | -0.033 |
| ushbp1 | 0.529 | mrpl57 | -0.033 |