PDS5 cohesin associated factor A
ZFIN 
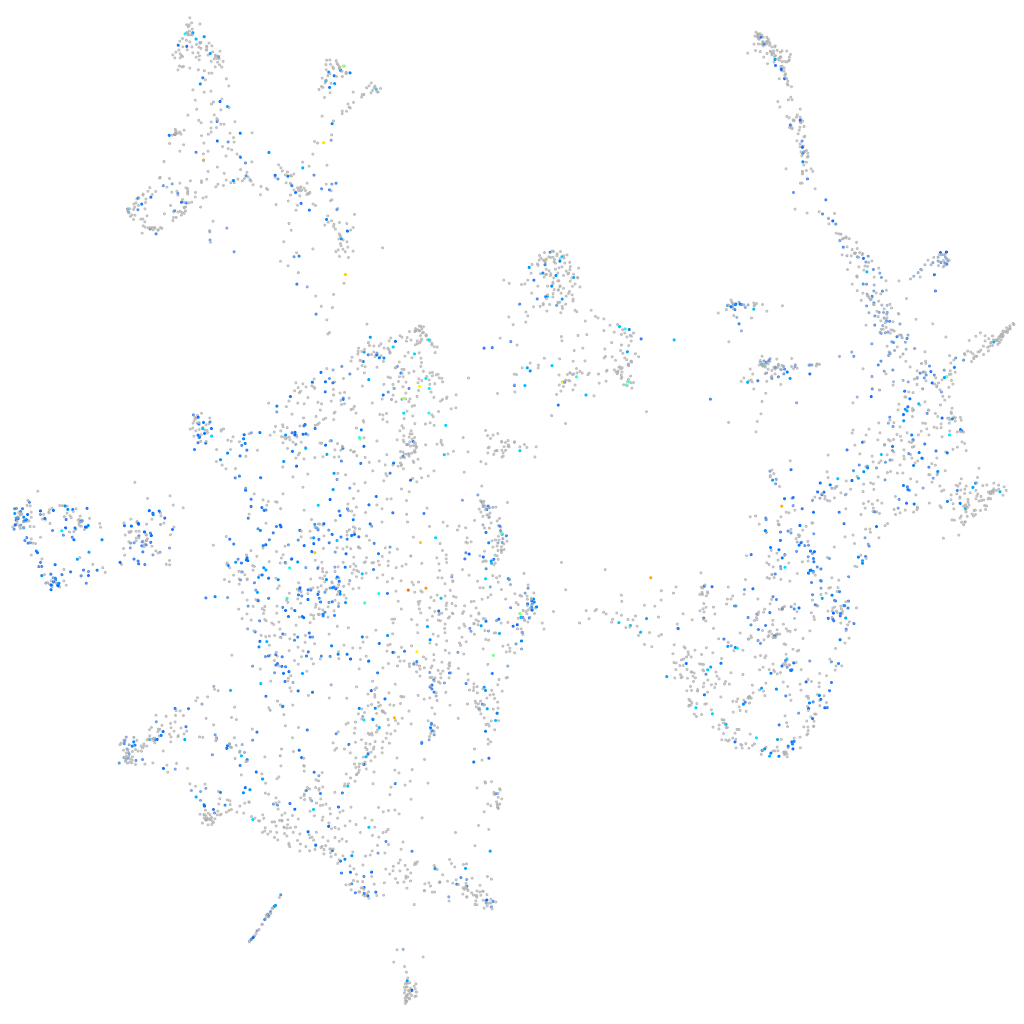
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| seta | 0.169 | pvalb2 | -0.091 |
| hmgb2a | 0.166 | atp2b1a | -0.090 |
| ranbp1 | 0.166 | tob1b | -0.084 |
| snrpg | 0.166 | vamp2 | -0.082 |
| hnrnpa0l | 0.161 | s100t | -0.081 |
| snrpf | 0.159 | rnasekb | -0.080 |
| setb | 0.158 | nupr1a | -0.079 |
| hmgb2b | 0.157 | kif1aa | -0.078 |
| ddx39ab | 0.152 | apoa1b | -0.076 |
| snrpb | 0.151 | eno1a | -0.075 |
| pcna | 0.151 | anxa5a | -0.074 |
| h2afvb | 0.149 | calml4a | -0.074 |
| stmn1a | 0.149 | otofb | -0.071 |
| hmga1a | 0.148 | si:dkey-16p21.8 | -0.070 |
| h2afva | 0.148 | osbpl1a | -0.069 |
| si:ch211-222l21.1 | 0.146 | cpn1 | -0.068 |
| apex1 | 0.146 | col11a2 | -0.068 |
| marcksb | 0.146 | gb:bc139872 | -0.067 |
| hnrnpabb | 0.145 | ucmab | -0.067 |
| mki67 | 0.143 | pvalb8 | -0.067 |
| snrpd2 | 0.143 | slc17a8 | -0.067 |
| ccna2 | 0.142 | gapdhs | -0.066 |
| h2afx | 0.141 | ip6k2a | -0.066 |
| khdrbs1a | 0.141 | si:ch211-255p10.3 | -0.066 |
| rrm1 | 0.141 | calm1a | -0.066 |
| ran | 0.140 | tmem178b | -0.066 |
| hmgn2 | 0.140 | gpx2 | -0.065 |
| hmgb1b | 0.140 | apoa2 | -0.065 |
| dut | 0.140 | actc1b | -0.065 |
| h3f3d | 0.139 | loxhd1b | -0.065 |
| nucks1a | 0.138 | si:ch211-57i17.5 | -0.065 |
| rpa3 | 0.138 | apba1a | -0.065 |
| anp32a | 0.137 | dpysl3 | -0.065 |
| hnrnpa0b | 0.137 | clic5b | -0.065 |
| rbbp4 | 0.136 | tmbim1b | -0.065 |