PDZ and LIM domain 1 (elfin)
ZFIN 
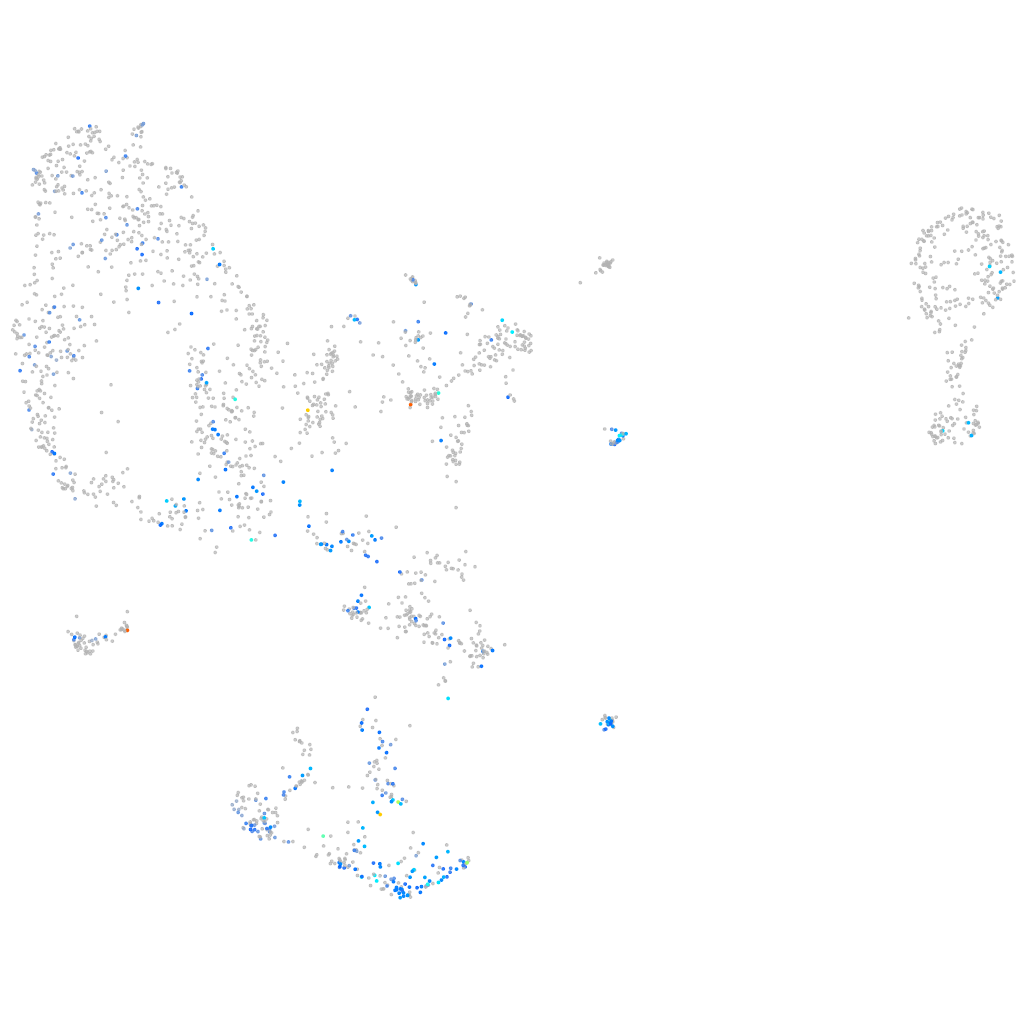
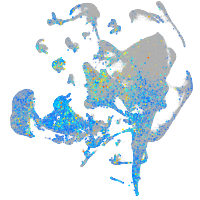
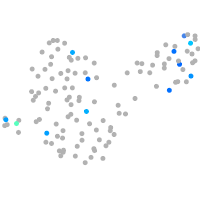
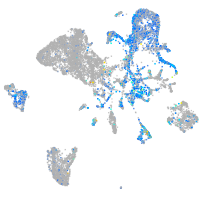
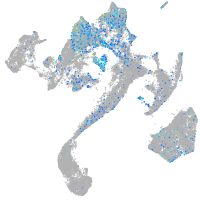
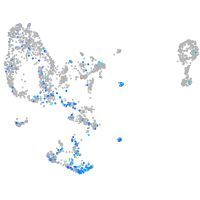
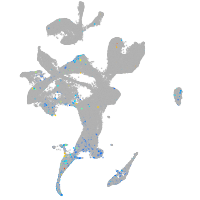
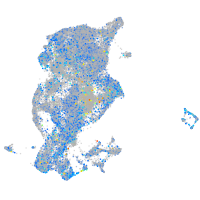
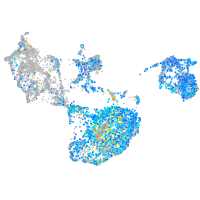
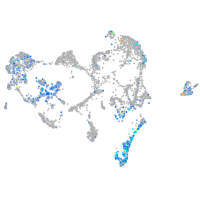
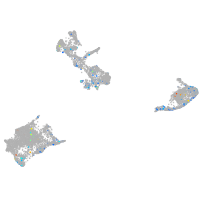
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| ctgfa | 0.324 | grhprb | -0.150 |
| nfkbiaa | 0.311 | acy3.2 | -0.146 |
| rnf183 | 0.309 | hao1 | -0.134 |
| elf3 | 0.308 | hoga1 | -0.131 |
| phlda2 | 0.302 | dab2 | -0.129 |
| tagln2 | 0.297 | pnp6 | -0.128 |
| atp1a1a.3 | 0.295 | zgc:136493 | -0.128 |
| ier2b | 0.295 | si:dkey-33i11.9 | -0.126 |
| vsig8b | 0.292 | pdzk1ip1 | -0.125 |
| ahnak | 0.291 | si:dkey-194e6.1 | -0.124 |
| gdpd3a | 0.289 | upb1 | -0.124 |
| si:dkey-36i7.3 | 0.289 | ftcd | -0.123 |
| soul2 | 0.283 | fbp1b | -0.121 |
| thbs1b | 0.281 | nit2 | -0.119 |
| prkag2a | 0.279 | dpys | -0.119 |
| gpa33a | 0.279 | pklr | -0.118 |
| agr2 | 0.278 | slc26a1 | -0.117 |
| zgc:158423 | 0.277 | smim1 | -0.116 |
| gadd45bb | 0.275 | phyhd1 | -0.116 |
| ccl19a.1 | 0.275 | gpt2l | -0.116 |
| zgc:64022 | 0.273 | gstt1a | -0.114 |
| trim35-12 | 0.272 | dpydb | -0.112 |
| tbx2b | 0.271 | si:dkey-28n18.9 | -0.112 |
| ppp1r1b | 0.271 | cx32.3 | -0.112 |
| mal2 | 0.270 | gamt | -0.112 |
| myh9a | 0.269 | wu:fb97g03 | -0.111 |
| prrg2 | 0.268 | alpl | -0.111 |
| f2rl1.2 | 0.265 | cubn | -0.111 |
| si:ch73-335l21.4 | 0.265 | zgc:162816 | -0.111 |
| chac1 | 0.263 | xirp1 | -0.109 |
| krt8 | 0.263 | epdl2 | -0.108 |
| sema3fa | 0.262 | nipsnap3a | -0.108 |
| junba | 0.260 | lgmn | -0.108 |
| clcnk | 0.260 | ncl | -0.108 |
| bsnd | 0.259 | snrnp70 | -0.107 |