"phosphodiesterase 3A, cGMP-inhibited"
ZFIN 
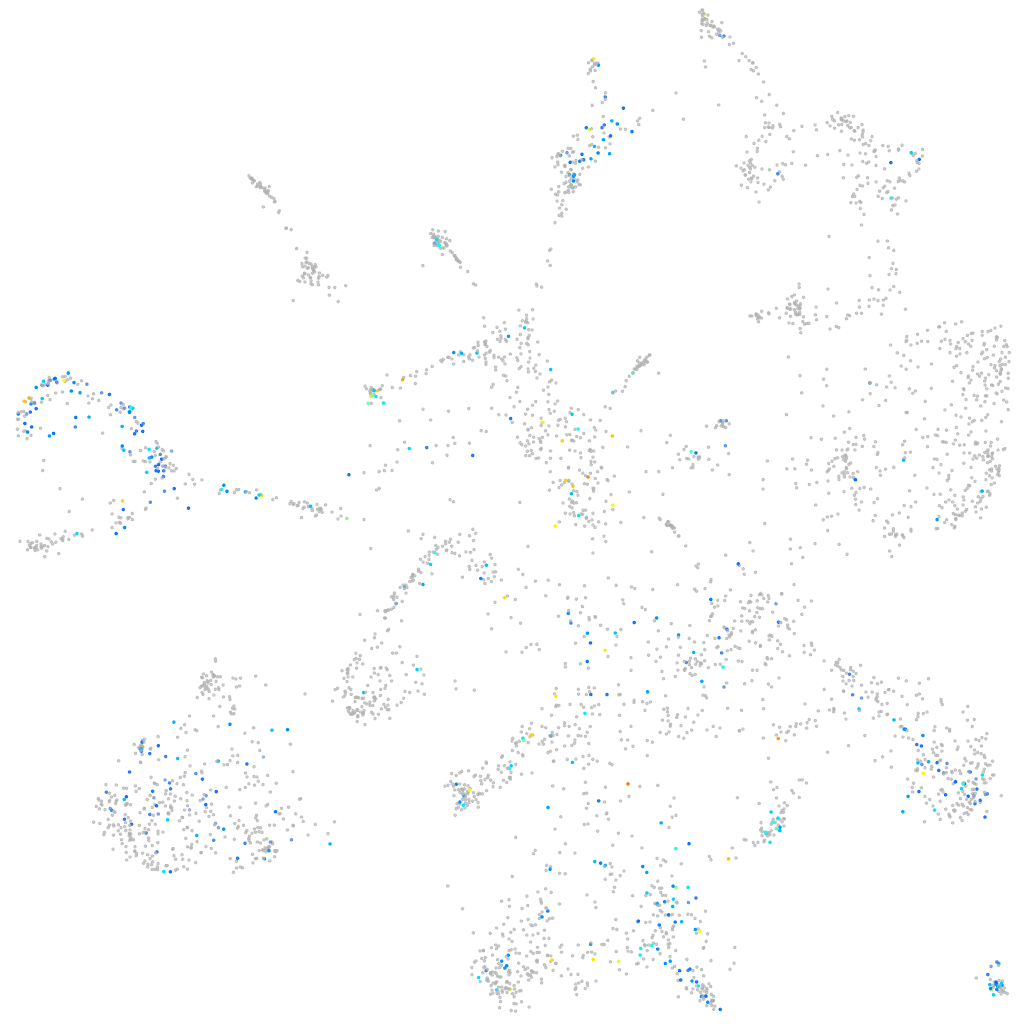
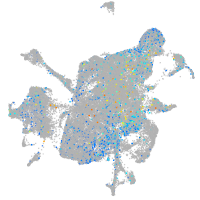
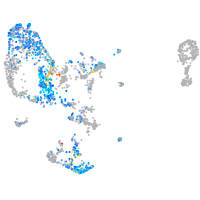
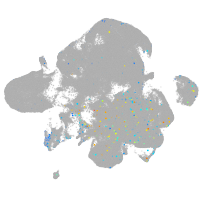
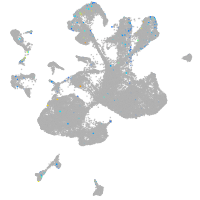
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| gucy1a1 | 0.170 | jam2b | -0.136 |
| pfkpa | 0.165 | podxl | -0.124 |
| slc16a3 | 0.164 | mdka | -0.104 |
| pgam1a | 0.161 | tmem88b | -0.100 |
| fbp2 | 0.157 | aqp1a.1 | -0.097 |
| gapdhs | 0.153 | COLEC10 | -0.093 |
| acta2 | 0.152 | krt94 | -0.092 |
| pkma | 0.151 | si:ch211-39i22.1 | -0.085 |
| lmod1b | 0.150 | hspb1 | -0.084 |
| tpm1 | 0.149 | slc43a3b | -0.083 |
| aldocb | 0.149 | anxa1a | -0.082 |
| chadlb | 0.147 | s100a10b | -0.082 |
| ldha | 0.145 | rhag | -0.082 |
| ak1 | 0.143 | colec11 | -0.082 |
| gucy1b1 | 0.142 | rbp4 | -0.081 |
| myl9a | 0.141 | tgm2b | -0.080 |
| pgk1 | 0.141 | krt18b | -0.079 |
| tagln | 0.140 | cd151 | -0.078 |
| mylkb | 0.139 | cx43.4 | -0.075 |
| eno3 | 0.136 | wt1b | -0.075 |
| slc2a1b | 0.135 | rps12 | -0.074 |
| akap7 | 0.134 | si:ch211-156j16.1 | -0.074 |
| eno1a | 0.134 | naca | -0.074 |
| cnn1a | 0.132 | c1qtnf6a | -0.074 |
| casz1 | 0.131 | tspan18a | -0.073 |
| tpm2 | 0.130 | ftr82 | -0.072 |
| cx43 | 0.130 | gstm.3 | -0.071 |
| wnt11r | 0.129 | krt8 | -0.070 |
| si:ch73-139j3.4 | 0.127 | rpl27 | -0.069 |
| si:ch211-270g19.5 | 0.126 | eppk1 | -0.069 |
| BX088707.3 | 0.125 | palm1b | -0.069 |
| myh11a | 0.125 | col18a1a | -0.068 |
| crispld2 | 0.124 | krt15 | -0.068 |
| hk1 | 0.122 | cdon | -0.068 |
| actn1 | 0.121 | marcksl1b | -0.068 |