piccolo presynaptic cytomatrix protein b
ZFIN 
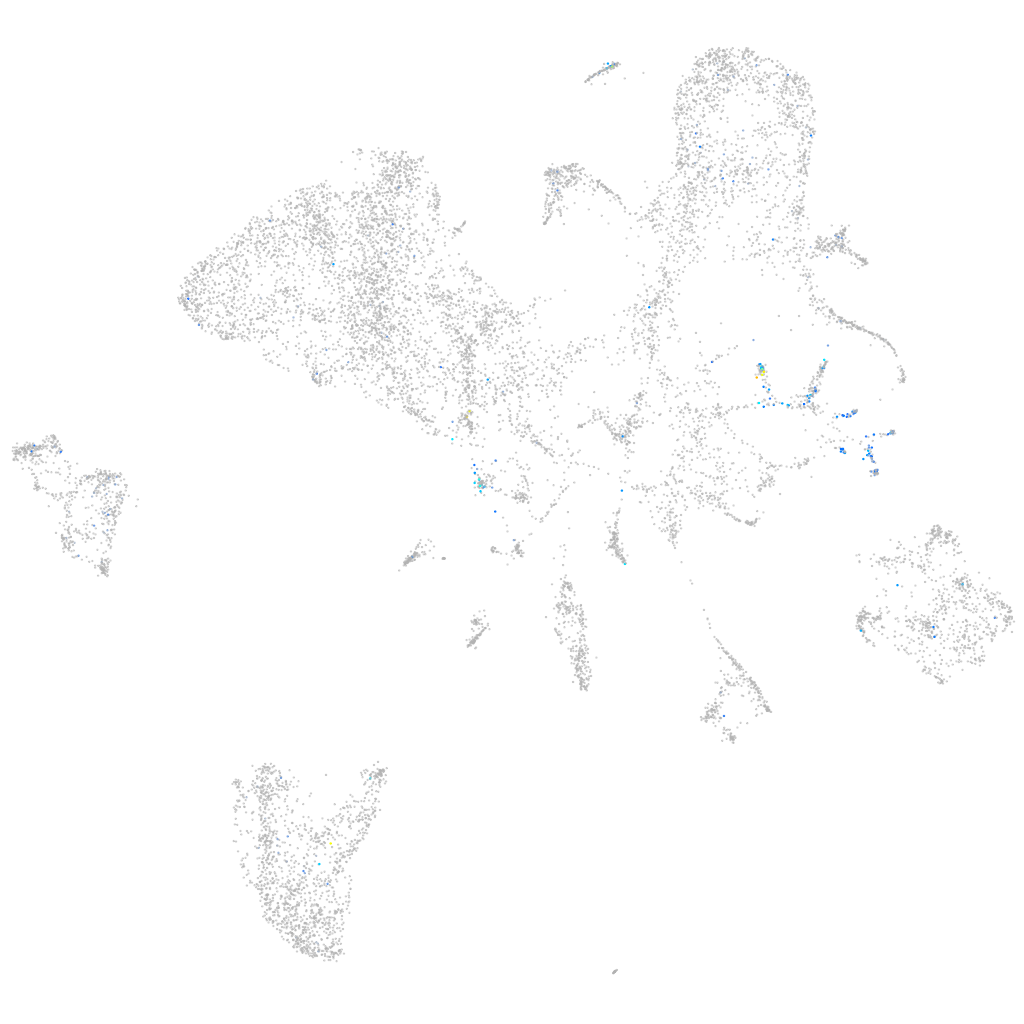
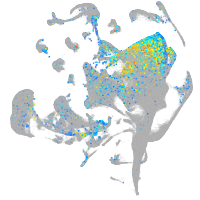
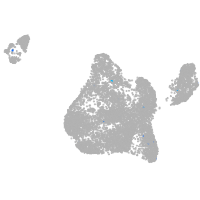
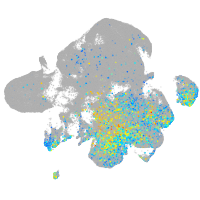
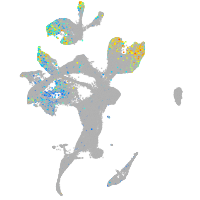
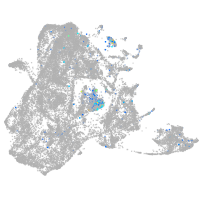
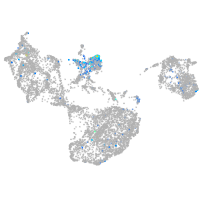
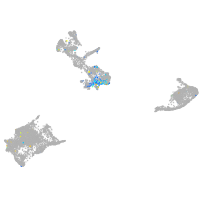
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| scg3 | 0.277 | ahcy | -0.096 |
| pcsk1nl | 0.261 | gapdh | -0.088 |
| vamp2 | 0.254 | gamt | -0.086 |
| vat1 | 0.251 | rps20 | -0.083 |
| atp6v0cb | 0.249 | aldob | -0.080 |
| neurod1 | 0.248 | nupr1b | -0.078 |
| scg2b | 0.245 | gatm | -0.077 |
| aldocb | 0.242 | rpl6 | -0.075 |
| scn3b | 0.241 | hdlbpa | -0.074 |
| arf3a | 0.238 | mat1a | -0.072 |
| cpe | 0.235 | gstt1a | -0.072 |
| syt1a | 0.233 | hspe1 | -0.071 |
| pax6b | 0.232 | fbp1b | -0.071 |
| mir7a-1 | 0.230 | bhmt | -0.069 |
| tspan7b | 0.228 | aldh6a1 | -0.069 |
| scgn | 0.228 | gstr | -0.068 |
| rims2a | 0.223 | apoa4b.1 | -0.067 |
| pcsk2 | 0.222 | aqp12 | -0.067 |
| si:dkey-153k10.9 | 0.221 | gnmt | -0.066 |
| pcsk1 | 0.216 | apoc2 | -0.066 |
| scg2a | 0.214 | aldh7a1 | -0.066 |
| XLOC-026746 | 0.211 | mgst1.2 | -0.066 |
| cacna1c | 0.211 | cx32.3 | -0.066 |
| insm1b | 0.208 | si:dkey-16p21.8 | -0.066 |
| si:dkey-280e21.3 | 0.206 | rgrb | -0.066 |
| atpv0e2 | 0.206 | aldh9a1a.1 | -0.066 |
| prlra | 0.204 | agxtb | -0.065 |
| cabp5b | 0.204 | dhdhl | -0.065 |
| rapgef4 | 0.203 | apoc1 | -0.064 |
| kcnj11 | 0.202 | scp2a | -0.064 |
| gnao1a | 0.201 | eno3 | -0.063 |
| kcnc3a | 0.200 | shmt1 | -0.063 |
| rnasekb | 0.198 | grhprb | -0.063 |
| egr4 | 0.198 | cebpd | -0.063 |
| cplx2l | 0.197 | wu:fj16a03 | -0.062 |