protocadherin 2 alpha b2
ZFIN 
Other cell groups


all cells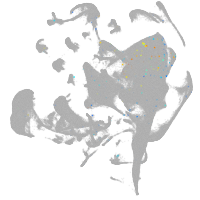 |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme 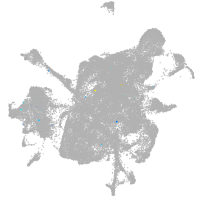 |
hematopoietic / vasculature 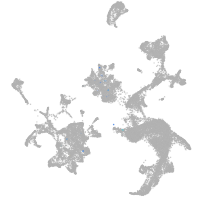 |
pronephros 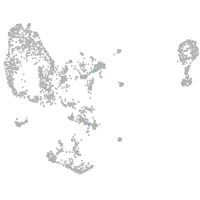 |
neural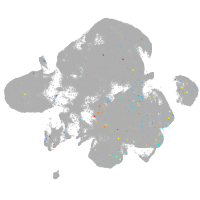 |
spinal cord / glia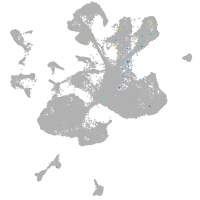 |
eye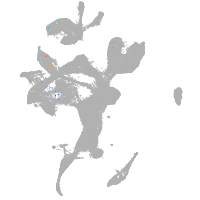 |
taste / olfactory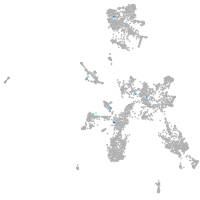 |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| elavl3 | 0.061 | aldob | -0.028 |
| tubb5 | 0.058 | ccng1 | -0.023 |
| stmn1b | 0.056 | id1 | -0.023 |
| gng3 | 0.055 | eef1da | -0.022 |
| rtn1a | 0.054 | eif4ebp3l | -0.022 |
| tuba1c | 0.054 | hdlbpa | -0.022 |
| mllt11 | 0.052 | npm1a | -0.022 |
| tmsb | 0.052 | vamp3 | -0.022 |
| rtn1b | 0.051 | atp1b1a | -0.021 |
| tmeff1b | 0.051 | banf1 | -0.021 |
| zc4h2 | 0.051 | cfl1l | -0.021 |
| si:dkey-276j7.1 | 0.050 | dkc1 | -0.021 |
| ywhah | 0.050 | epcam | -0.021 |
| gap43 | 0.049 | hspb1 | -0.021 |
| gng2 | 0.049 | rsl1d1 | -0.021 |
| hmgb3a | 0.049 | tuba8l | -0.021 |
| nova2 | 0.049 | bzw1b | -0.020 |
| dpysl3 | 0.048 | ccnd1 | -0.020 |
| fez1 | 0.047 | chaf1a | -0.020 |
| gpm6ab | 0.047 | fbl | -0.020 |
| myt1a | 0.047 | nop10 | -0.020 |
| myt1b | 0.047 | nop58 | -0.020 |
| elavl4 | 0.046 | tuba8l2 | -0.020 |
| stmn2a | 0.046 | apoeb | -0.019 |
| vamp2 | 0.046 | f11r.1 | -0.019 |
| sncb | 0.045 | fosab | -0.019 |
| dpysl2b | 0.045 | FQ323156.1 | -0.019 |
| gpm6aa | 0.044 | lig1 | -0.019 |
| tuba1a | 0.044 | nutf2l | -0.019 |
| ywhag2 | 0.044 | pcna | -0.019 |
| stxbp1a | 0.044 | pfn1 | -0.019 |
| adam22 | 0.043 | sdc4 | -0.019 |
| atp6v0cb | 0.043 | cd9b | -0.018 |
| cnrip1a | 0.043 | gsta.1 | -0.018 |
| jpt1b | 0.043 | gstp1 | -0.018 |




