protocadherin 20
ZFIN 
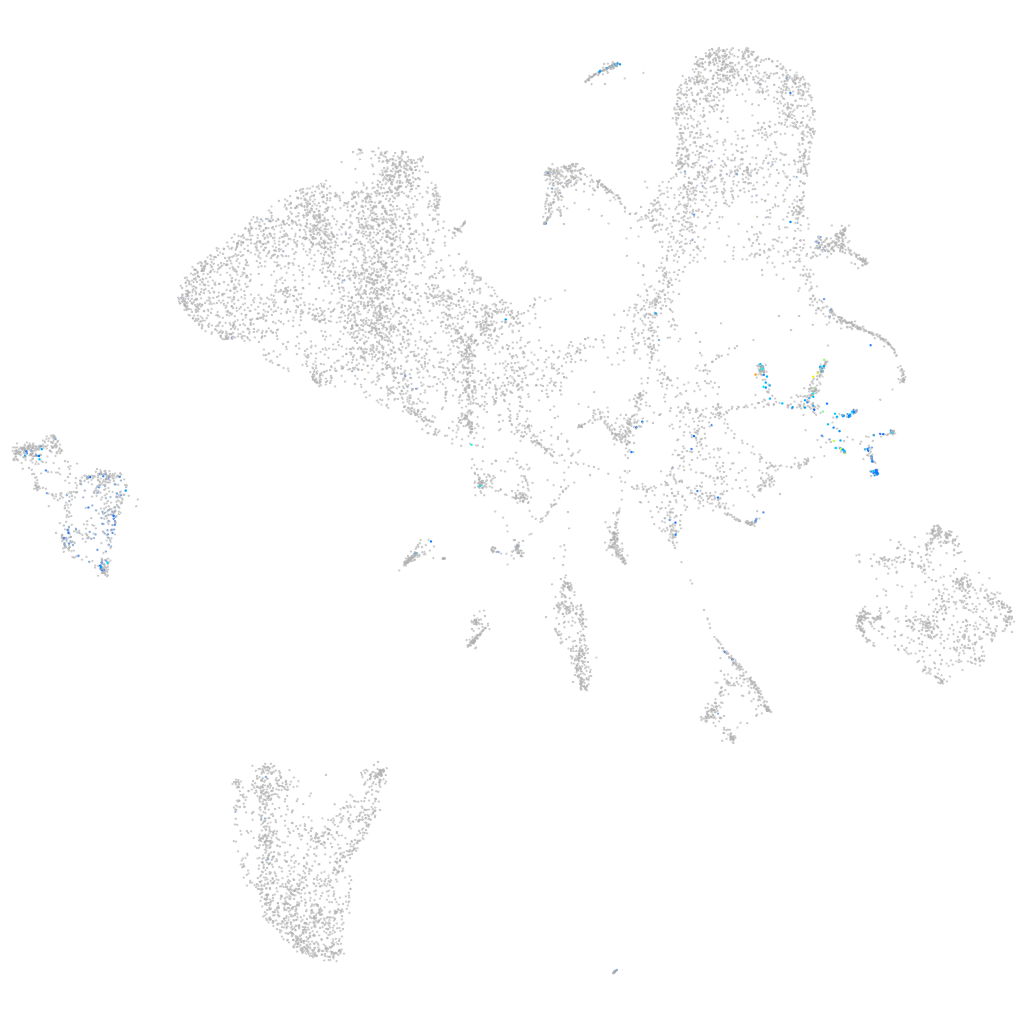
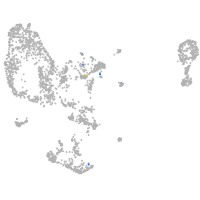
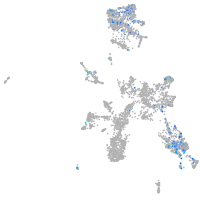
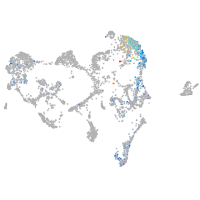
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| scg3 | 0.370 | gapdh | -0.151 |
| pax6b | 0.353 | gamt | -0.138 |
| mir7a-1 | 0.346 | ahcy | -0.127 |
| si:dkey-153k10.9 | 0.344 | gatm | -0.124 |
| neurod1 | 0.341 | bhmt | -0.118 |
| scgn | 0.337 | mat1a | -0.113 |
| pcsk1nl | 0.333 | fbp1b | -0.112 |
| pcsk2 | 0.320 | apoa4b.1 | -0.112 |
| syt1a | 0.315 | apoc2 | -0.110 |
| rprmb | 0.314 | apoa1b | -0.109 |
| scg2a | 0.311 | cx32.3 | -0.109 |
| vat1 | 0.309 | gstt1a | -0.106 |
| kcnj11 | 0.303 | afp4 | -0.106 |
| rasd4 | 0.300 | hspe1 | -0.105 |
| pcsk1 | 0.300 | apoc1 | -0.105 |
| ndufa4l2a | 0.289 | aqp12 | -0.104 |
| SLC5A10 | 0.286 | nupr1b | -0.102 |
| trhra | 0.283 | agxtb | -0.100 |
| isl1 | 0.277 | agxta | -0.099 |
| rims2a | 0.271 | gnmt | -0.099 |
| abhd15a | 0.269 | glud1b | -0.099 |
| jph3 | 0.265 | si:dkey-16p21.8 | -0.098 |
| id4 | 0.263 | aldh6a1 | -0.098 |
| insm1a | 0.262 | scp2a | -0.097 |
| spon1b | 0.259 | gstr | -0.096 |
| rem2 | 0.259 | apoa2 | -0.095 |
| tspan7b | 0.259 | aldh7a1 | -0.095 |
| pygma | 0.258 | apobb.1 | -0.095 |
| camk1da | 0.255 | aldob | -0.095 |
| gapdhs | 0.253 | tfa | -0.094 |
| insm1b | 0.252 | pnp4b | -0.093 |
| gpc1a | 0.252 | gcshb | -0.092 |
| foxo1a | 0.251 | shmt1 | -0.092 |
| egr4 | 0.250 | eno3 | -0.089 |
| ins | 0.249 | serpina1l | -0.089 |