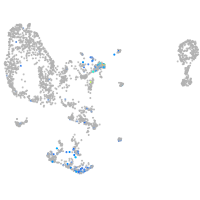
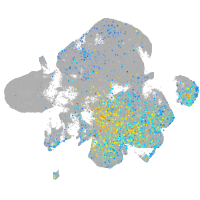
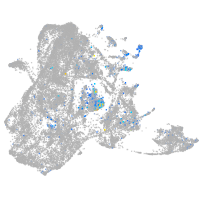
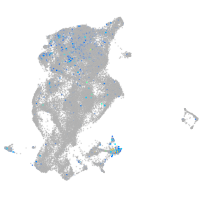
protocadherin 11
ZFIN 
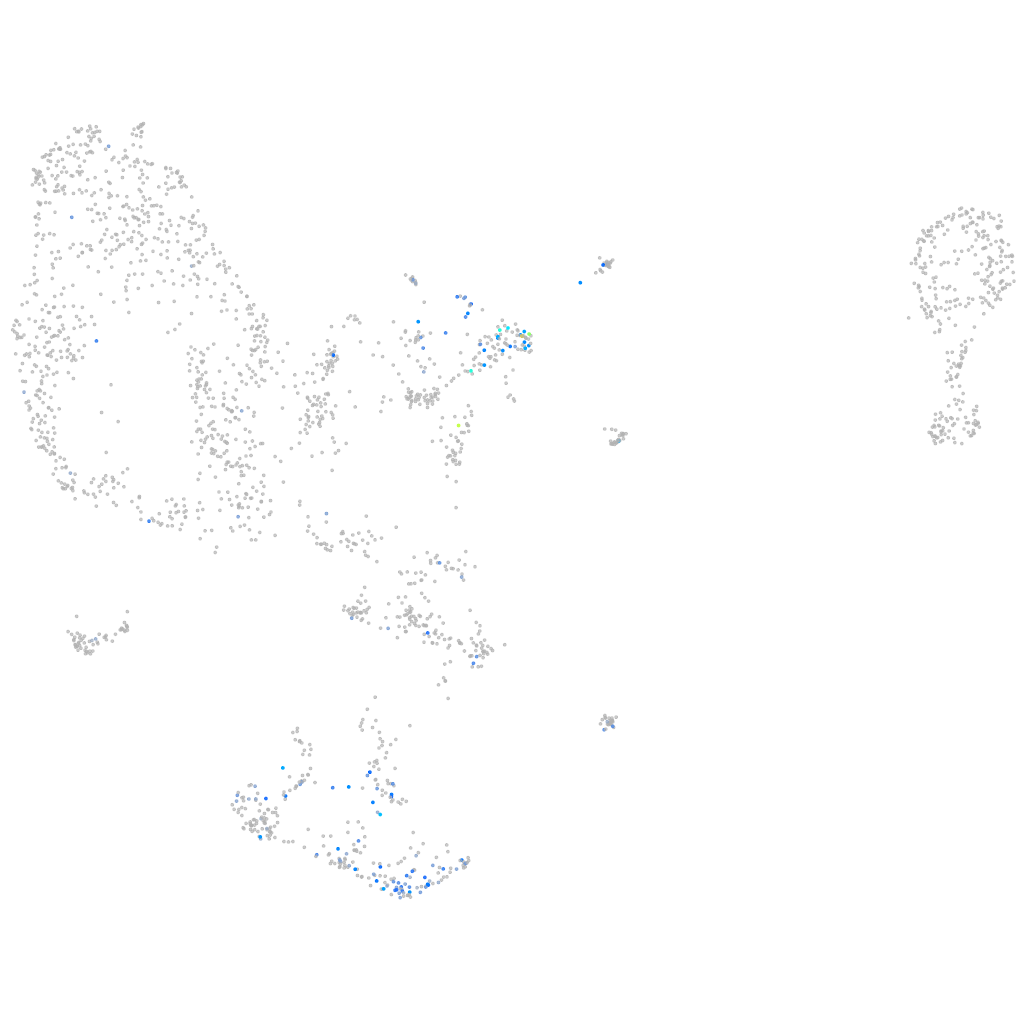
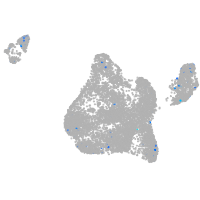
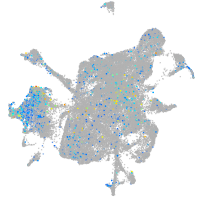
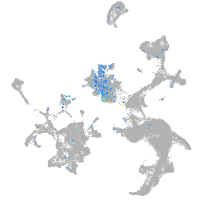
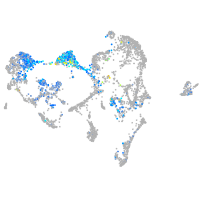
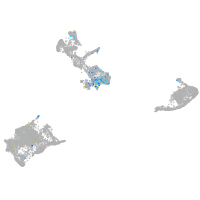
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| cacng2a | 0.363 | si:dkey-151g10.6 | -0.153 |
| DPYSL2 (1 of many) | 0.330 | acy3.2 | -0.140 |
| cabp7b | 0.330 | ezra | -0.124 |
| srgap1b | 0.322 | si:rp71-17i16.6 | -0.120 |
| gng3 | 0.322 | rplp1 | -0.116 |
| gpm6aa | 0.317 | ftcd | -0.115 |
| fez1 | 0.316 | eif5a2 | -0.114 |
| zgc:153426 | 0.315 | add3a | -0.114 |
| XLOC-011956 | 0.302 | adka | -0.112 |
| aplp1 | 0.300 | npm1a | -0.110 |
| prelid3a | 0.298 | lrp2a | -0.110 |
| stxbp1a | 0.291 | si:dkey-194e6.1 | -0.110 |
| uchl1 | 0.281 | cubn | -0.110 |
| map1aa | 0.280 | pnp6 | -0.110 |
| mras | 0.279 | pdzk1ip1 | -0.109 |
| ywhag2 | 0.277 | slc26a6 | -0.109 |
| sypa | 0.277 | npr1b | -0.109 |
| cpne8 | 0.275 | fmo5 | -0.108 |
| gnao1b | 0.275 | slc9a3r1a | -0.108 |
| malrd1 | 0.274 | alpl | -0.107 |
| cadm3 | 0.274 | pdzk1 | -0.106 |
| smarcc2 | 0.272 | gnpda1 | -0.106 |
| stmn1b | 0.271 | serbp1a | -0.106 |
| si:ch211-215p11.1 | 0.270 | rps28 | -0.106 |
| tmem240b | 0.269 | rpl18 | -0.105 |
| ache | 0.266 | hoga1 | -0.105 |
| khdrbs2 | 0.266 | rps12 | -0.104 |
| grin1b | 0.266 | rpl34 | -0.104 |
| caly | 0.265 | slc26a1 | -0.103 |
| amph | 0.264 | ppiaa | -0.103 |
| id4 | 0.264 | eef1a1l1 | -0.102 |
| atp6v0a1b | 0.263 | dab2 | -0.102 |
| gapdhs | 0.261 | gamt | -0.102 |
| atp2b3a | 0.261 | slc4a4a | -0.102 |
| ppfia4 | 0.260 | fbp1b | -0.101 |