pterin-4 alpha-carbinolamine dehydratase/dimerization cofactor of hepatocyte nuclear factor 1 alpha
ZFIN 
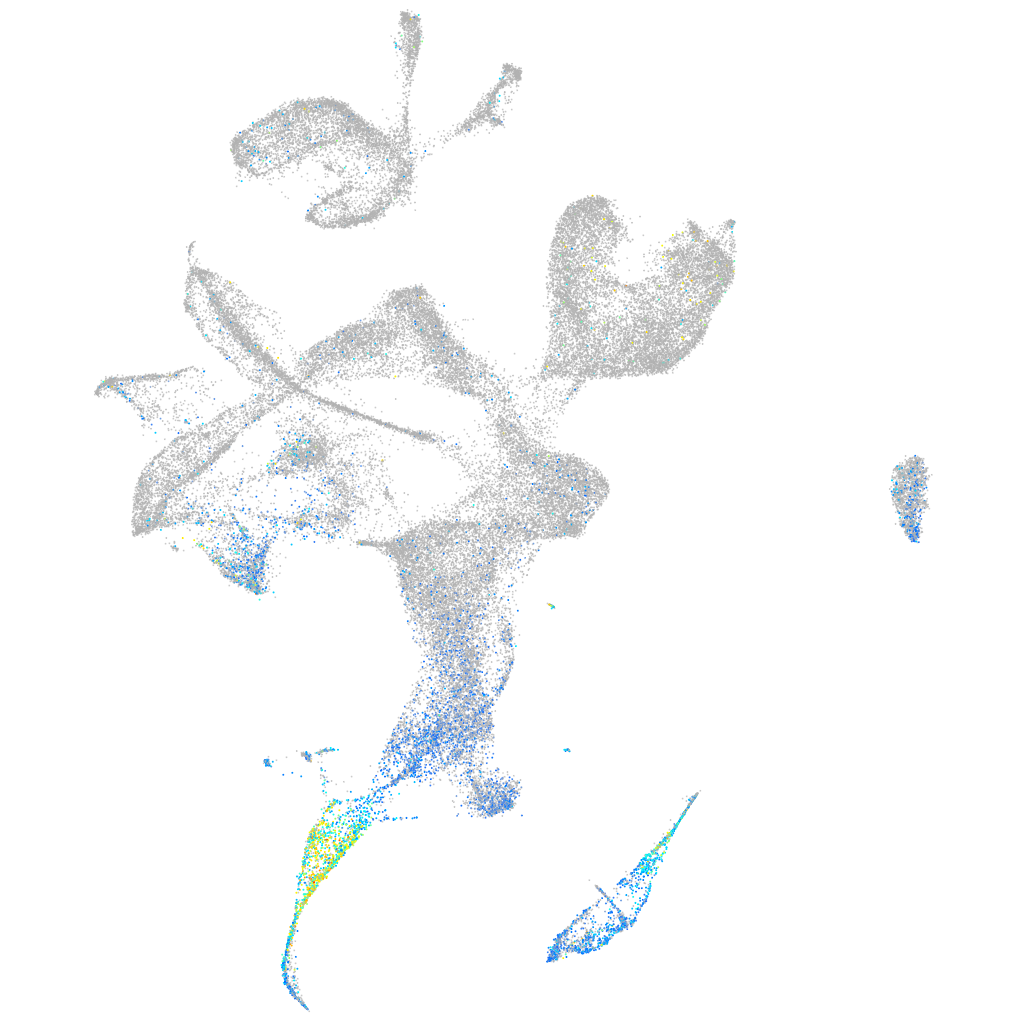
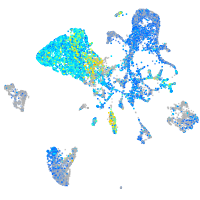
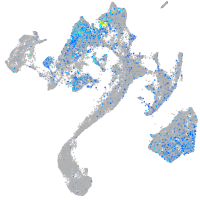
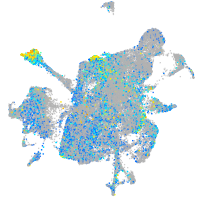
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| tspan36 | 0.559 | ptmab | -0.372 |
| slc45a2 | 0.548 | si:ch73-1a9.3 | -0.263 |
| dct | 0.544 | ptmaa | -0.252 |
| qdpra | 0.536 | hmgn6 | -0.236 |
| gpr143 | 0.527 | gpm6aa | -0.229 |
| fabp11a | 0.527 | tuba1c | -0.221 |
| pmela | 0.525 | hmgb1a | -0.216 |
| tyrp1b | 0.513 | ckbb | -0.192 |
| tyrp1a | 0.492 | cadm3 | -0.183 |
| rab38 | 0.491 | si:ch211-137a8.4 | -0.182 |
| pmelb | 0.484 | gpm6ab | -0.173 |
| zgc:110591 | 0.477 | CR383676.1 | -0.168 |
| mitfa | 0.477 | fabp7a | -0.166 |
| agtrap | 0.475 | tuba1a | -0.164 |
| tspan10 | 0.473 | marcksl1b | -0.162 |
| gstp1 | 0.472 | h2afva | -0.162 |
| sytl2b | 0.449 | epb41a | -0.161 |
| col4a5 | 0.444 | nova2 | -0.159 |
| oca2 | 0.443 | foxg1b | -0.157 |
| tmem88b | 0.440 | rorb | -0.156 |
| rab32a | 0.437 | anp32e | -0.154 |
| tyr | 0.436 | si:ch1073-429i10.3.1 | -0.150 |
| bace2 | 0.432 | snap25b | -0.148 |
| slc24a5 | 0.431 | btg1 | -0.147 |
| col4a6 | 0.427 | hmgn2 | -0.143 |
| sparc | 0.423 | neurod4 | -0.140 |
| fabp11b | 0.419 | stmn1b | -0.138 |
| FP085398.1 | 0.412 | mdkb | -0.137 |
| cracr2ab | 0.411 | h3f3c | -0.137 |
| pnp4a | 0.411 | rtn1a | -0.136 |
| cx43 | 0.404 | crx | -0.136 |
| rnaseka | 0.398 | hmgb1b | -0.136 |
| triobpa | 0.397 | rnasekb | -0.133 |
| pah | 0.383 | ndrg4 | -0.133 |
| trpm1b | 0.380 | hmgb3a | -0.129 |