poly (ADP-ribose) polymerase 2
ZFIN 
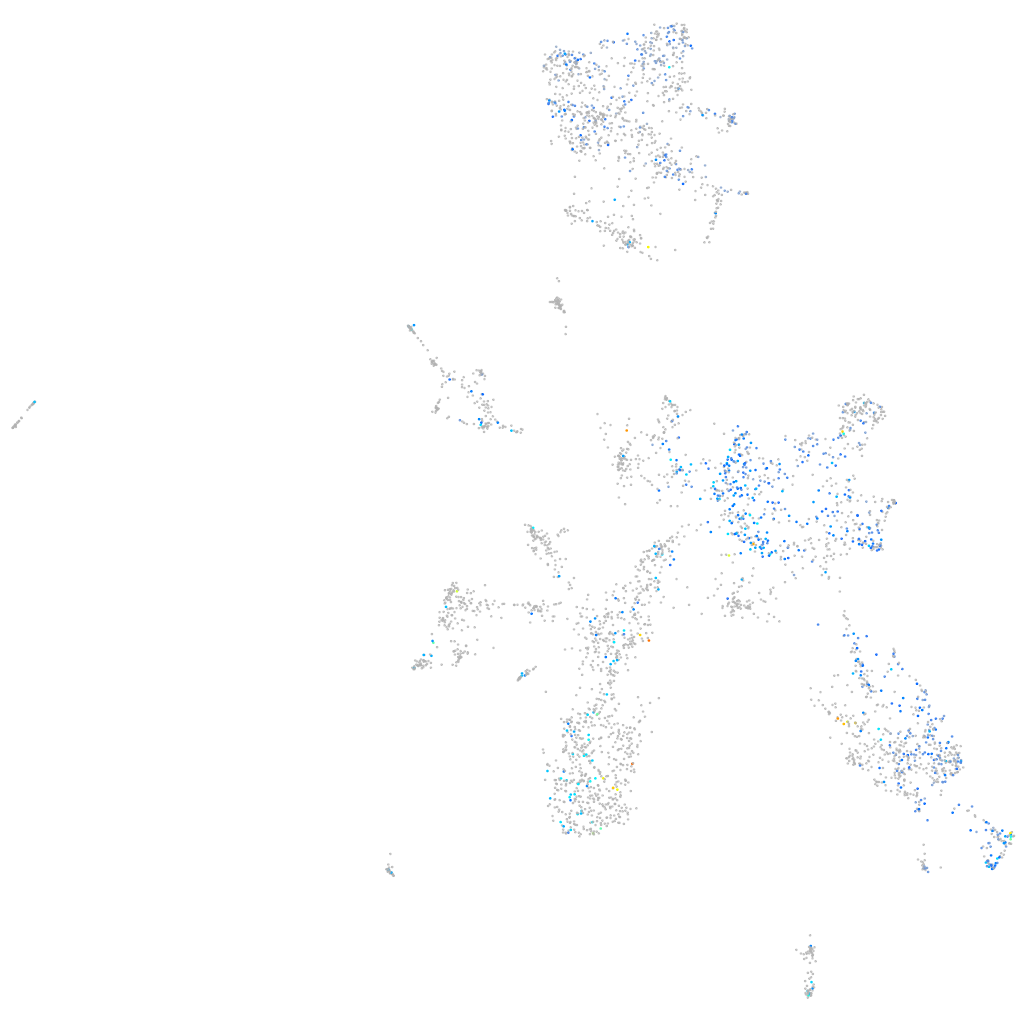
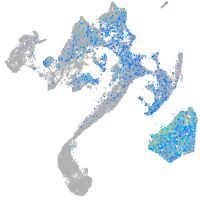
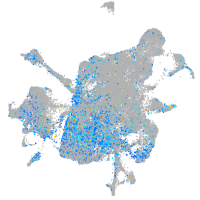
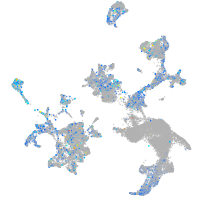
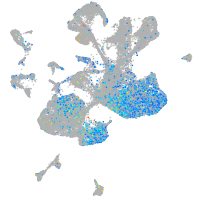
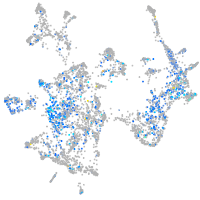
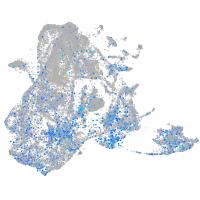
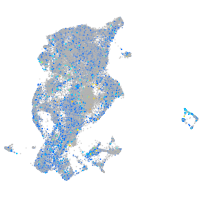
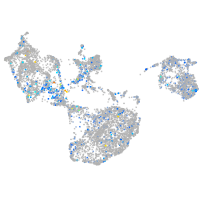
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| nasp | 0.228 | CR383676.1 | -0.134 |
| rrm1 | 0.226 | gabarapa | -0.102 |
| zgc:110540 | 0.217 | anxa5b | -0.102 |
| mibp | 0.214 | pvalb2 | -0.101 |
| si:ch211-156b7.4 | 0.213 | calm1a | -0.100 |
| rrm2 | 0.213 | pvalb1 | -0.098 |
| ncapg | 0.205 | smdt1b | -0.095 |
| dut | 0.204 | txn | -0.092 |
| rpa2 | 0.203 | scg2a | -0.092 |
| dek | 0.203 | syt1a | -0.091 |
| stmn1a | 0.201 | atp6v0cb | -0.088 |
| rbbp4 | 0.198 | vamp2 | -0.087 |
| mcm7 | 0.198 | xbp1 | -0.087 |
| chaf1a | 0.197 | calr | -0.087 |
| smc2 | 0.197 | bik | -0.085 |
| pcna | 0.194 | zgc:165573 | -0.084 |
| lbr | 0.194 | rtn1a | -0.084 |
| CABZ01005379.1 | 0.192 | calb2a | -0.083 |
| zgc:110216 | 0.191 | gng3 | -0.082 |
| cks1b | 0.191 | tmem54a | -0.082 |
| ccne2 | 0.189 | ppp3r1a | -0.081 |
| lig1 | 0.187 | zgc:65894 | -0.081 |
| her2 | 0.187 | gstp1 | -0.081 |
| nutf2l | 0.186 | gapdhs | -0.080 |
| slbp | 0.184 | h1f0 | -0.079 |
| cdca5 | 0.183 | tmem59 | -0.079 |
| tk1 | 0.183 | atp2b1b | -0.078 |
| dhfr | 0.182 | prr15la | -0.078 |
| XLOC-042899 | 0.181 | stxbp1a | -0.078 |
| primpol | 0.181 | rnasekb | -0.077 |
| ccna2 | 0.180 | carmil2 | -0.075 |
| fen1 | 0.177 | si:ch211-242b18.1 | -0.075 |
| mis12 | 0.176 | krt91 | -0.074 |
| suv39h1b | 0.176 | cpeb4b | -0.074 |
| LOC100330864 | 0.175 | rab2a | -0.073 |