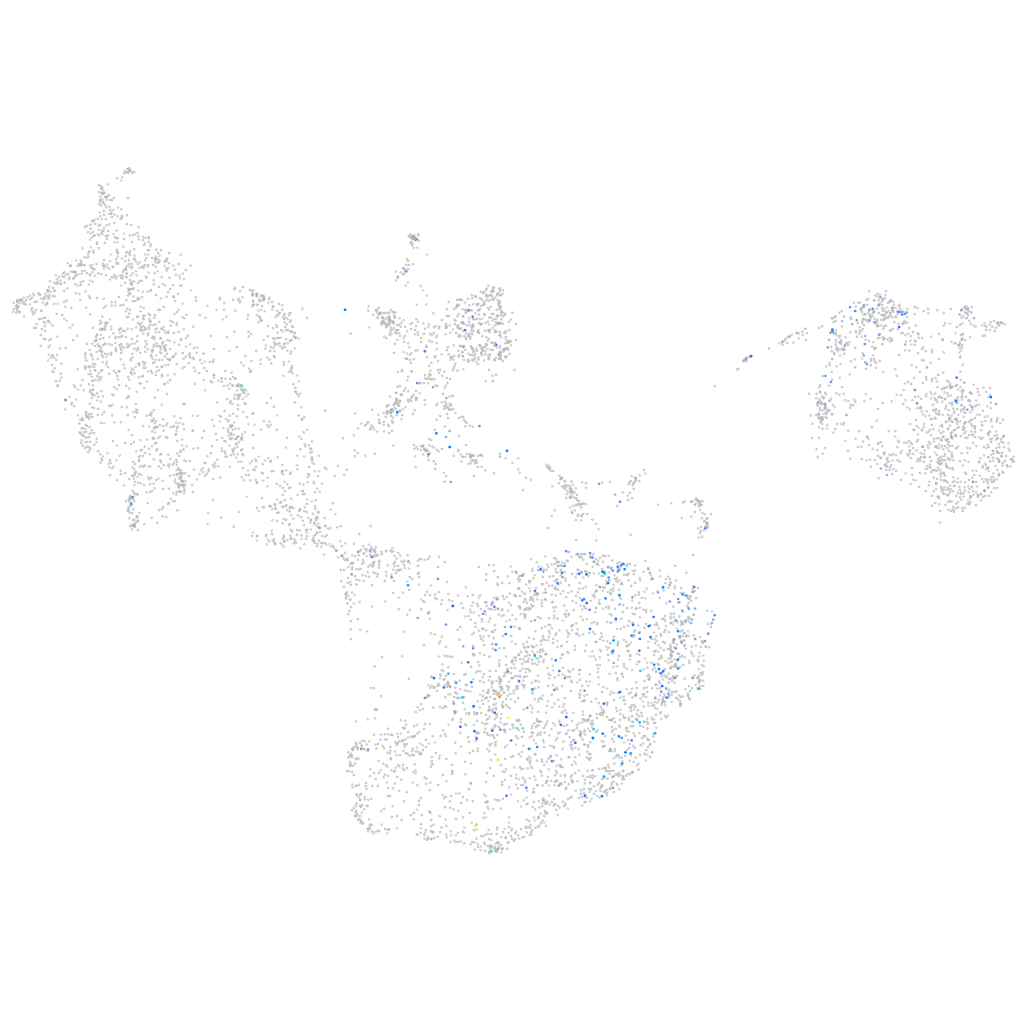
"purinergic receptor P2Y2, tandem duplicate 1"
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| LOC110438343 | 0.178 | marcksl1a | -0.116 |
| si:dkey-13n15.2 | 0.177 | c1qtnf5 | -0.106 |
| LOC108183599 | 0.170 | hgd | -0.100 |
| igfbp2a | 0.162 | fbln1 | -0.099 |
| si:ch211-119c20.2 | 0.157 | bhmt | -0.096 |
| col17a1a | 0.153 | ttc36 | -0.092 |
| krt5 | 0.153 | qdpra | -0.089 |
| zgc:85932 | 0.153 | cfl1 | -0.088 |
| cldn1 | 0.152 | cd82a | -0.087 |
| hcls1 | 0.152 | ptx3a | -0.086 |
| pfn1 | 0.149 | hpdb | -0.084 |
| cotl1 | 0.148 | prrx1a | -0.083 |
| ecrg4b | 0.144 | prrx1b | -0.083 |
| si:dkey-33c14.3 | 0.143 | pcbd1 | -0.082 |
| krt4 | 0.142 | twist1a | -0.082 |
| zgc:92380 | 0.142 | tmem119b | -0.081 |
| DSG2 | 0.142 | mfap2 | -0.080 |
| anxa1a | 0.141 | msna | -0.079 |
| cfl1l | 0.140 | gpc1a | -0.078 |
| krtt1c19e | 0.139 | pmp22a | -0.078 |
| zgc:77439 | 0.138 | cd81a | -0.077 |
| krt91 | 0.138 | fah | -0.076 |
| rbp4 | 0.137 | hoxa13b | -0.076 |
| col14a1b | 0.137 | pah | -0.074 |
| zdhhc21.1 | 0.137 | reck | -0.074 |
| plecb | 0.136 | twist3 | -0.073 |
| tmsb1 | 0.136 | calr | -0.073 |
| itga6b | 0.135 | tspan18b | -0.073 |
| si:dkey-71l4.2 | 0.135 | mdh1aa | -0.072 |
| col11a1a | 0.132 | stox1 | -0.072 |
| csrp1a | 0.132 | SCARF2 | -0.071 |
| CABZ01083546.1 | 0.132 | tuba8l3 | -0.071 |
| kera | 0.132 | gulp1a | -0.070 |
| cldni | 0.131 | slc16a10 | -0.070 |
| spaca4l | 0.130 | kazald3 | -0.070 |