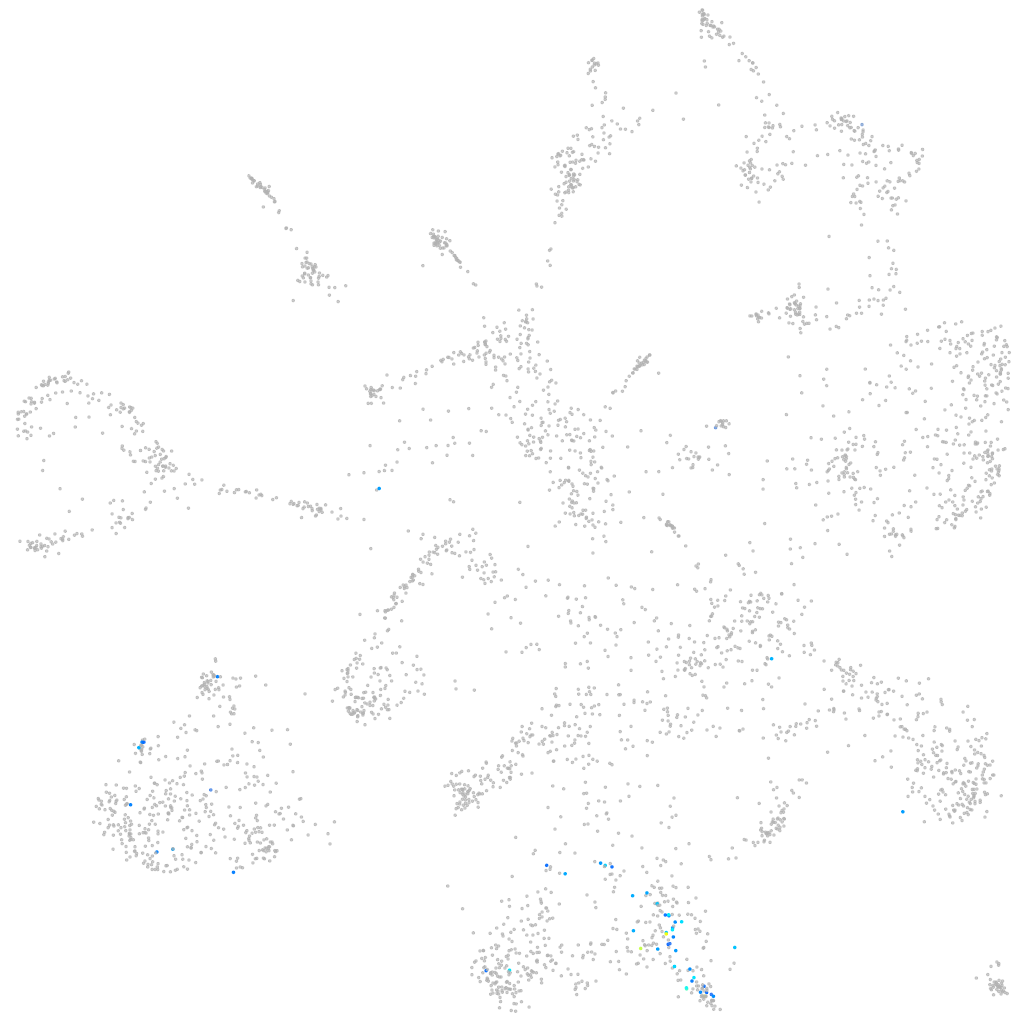
"purinergic receptor P2X, ligand-gated ion channel, 5"
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| actc1a | 0.392 | mdka | -0.089 |
| kcnk18 | 0.338 | si:dkey-261h17.1 | -0.075 |
| acta1b | 0.289 | krt8 | -0.071 |
| acta1a | 0.270 | krt18a.1 | -0.068 |
| desmb | 0.267 | krt94 | -0.068 |
| cald1b | 0.261 | pdgfra | -0.067 |
| cnn1b | 0.248 | fthl27 | -0.067 |
| cabp1a | 0.247 | CR383676.1 | -0.066 |
| csrp1b | 0.247 | bambia | -0.063 |
| pnp4a | 0.244 | marcksl1b | -0.062 |
| BX005329.2 | 0.243 | si:ch211-39i22.1 | -0.062 |
| CABZ01009512.2 | 0.243 | tmem98 | -0.061 |
| tpm2 | 0.239 | sparc | -0.060 |
| casz1 | 0.235 | fgfr2 | -0.060 |
| pdlim3b | 0.235 | krt18b | -0.059 |
| tspan15 | 0.227 | lsp1 | -0.058 |
| si:dkey-285b23.3 | 0.227 | jam2b | -0.056 |
| pdlim3a | 0.226 | rbpms2b | -0.055 |
| fbxl22 | 0.225 | ednraa | -0.054 |
| tuba8l2 | 0.221 | tmem88b | -0.054 |
| myh11a | 0.216 | podxl | -0.053 |
| prkd1 | 0.215 | nid2a | -0.053 |
| AL954694.1 | 0.214 | si:ch211-160j14.3 | -0.053 |
| leap2 | 0.204 | cd248a | -0.052 |
| arr3b | 0.202 | tmem176 | -0.052 |
| plk2b | 0.202 | fn1a | -0.051 |
| pgm5 | 0.202 | fbn2b | -0.050 |
| myl6 | 0.201 | si:ch1073-291c23.2 | -0.050 |
| tagln | 0.200 | nid1a | -0.049 |
| mylkb | 0.199 | notch3 | -0.049 |
| flnca | 0.199 | si:ch73-86n18.1 | -0.049 |
| vcla | 0.198 | ephb3 | -0.049 |
| rnf213b | 0.196 | si:ch73-281n10.2 | -0.048 |
| tpm1 | 0.194 | akap12b | -0.048 |
| itpka | 0.193 | col6a4a | -0.047 |