ornithine carbamoyltransferase
ZFIN 
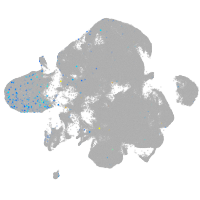
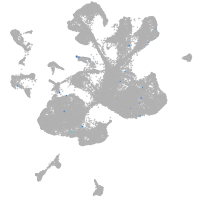
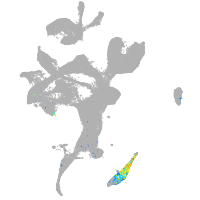
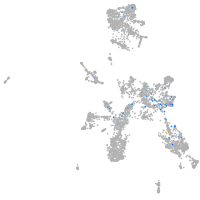
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| si:dkey-78a14.4 | 0.231 | nme2b.1 | -0.107 |
| PRRG3 | 0.207 | COX3 | -0.101 |
| si:ch211-247n2.1 | 0.207 | eef1da | -0.095 |
| LOC100147871 | 0.201 | rhbg | -0.093 |
| SLC6A13 | 0.196 | sod1 | -0.093 |
| slco1c1 | 0.182 | sod2 | -0.088 |
| myoz2a | 0.181 | rplp2 | -0.087 |
| prkg1a | 0.179 | mt-co2 | -0.085 |
| actr2b | 0.176 | mt-atp6 | -0.084 |
| amy2a | 0.172 | mt-co1 | -0.083 |
| p2ry13 | 0.160 | b2ml | -0.080 |
| itga2b | 0.160 | selenow2b | -0.076 |
| LOC101885690 | 0.160 | trim35-12 | -0.075 |
| XLOC-016965 | 0.158 | atp1a1a.3 | -0.075 |
| si:dkey-85k15.4 | 0.157 | rgs5b | -0.074 |
| si:ch73-111k22.3 | 0.155 | slc4a4b | -0.072 |
| XLOC-022751 | 0.154 | stat1a | -0.072 |
| stxbp6l | 0.154 | tdh | -0.071 |
| mir223 | 0.154 | pvalb1 | -0.070 |
| AL954142.1 | 0.153 | mt-nd2 | -0.069 |
| phactr1 | 0.152 | pvalb2 | -0.068 |
| fut8a | 0.150 | BX000438.2 | -0.068 |
| ptger1a | 0.149 | si:dkey-189h5.6 | -0.067 |
| htra4 | 0.148 | clcn2c | -0.066 |
| LOC101883456 | 0.147 | enosf1 | -0.065 |
| cmtm7 | 0.144 | ak1 | -0.065 |
| asb13a.1 | 0.143 | slc12a10.2 | -0.065 |
| hmga1a | 0.141 | tlcd1 | -0.065 |
| optc | 0.141 | zgc:162944 | -0.064 |
| fgl2a | 0.140 | rasd1 | -0.064 |
| mxtx2 | 0.140 | gsto1 | -0.064 |
| myoz1b | 0.139 | znf704 | -0.064 |
| krt92 | 0.138 | si:ch211-207n23.2 | -0.063 |
| susd1 | 0.136 | nfe2l2a | -0.063 |
| CR854907.1 | 0.136 | apoa1b | -0.063 |