oligosaccharyltransferase complex subunit 4 (non-catalytic)
ZFIN 
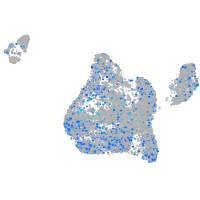
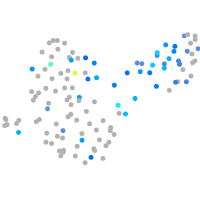
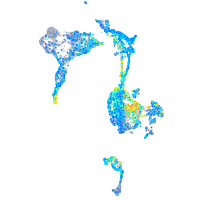
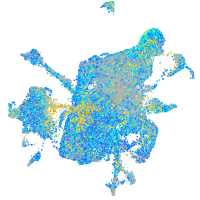
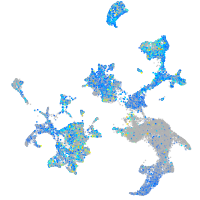
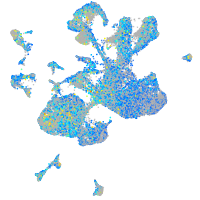
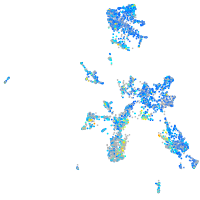
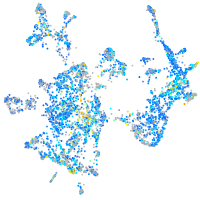
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| rpl38 | 0.187 | CR383676.1 | -0.157 |
| rps29 | 0.182 | gabarapb | -0.101 |
| rpl29 | 0.167 | rabl2 | -0.100 |
| rps28 | 0.153 | tspan13b | -0.096 |
| rpl37 | 0.147 | nptna | -0.095 |
| si:dkey-151g10.6 | 0.143 | histh1l | -0.094 |
| nme2b.1 | 0.142 | cep83 | -0.093 |
| metap2b | 0.137 | edem3 | -0.093 |
| icn | 0.135 | ompb | -0.092 |
| calr3b | 0.133 | dnal1 | -0.091 |
| s100a10b | 0.131 | map1aa | -0.090 |
| rpl39 | 0.131 | ctsd | -0.090 |
| cox7c | 0.130 | zgc:55461 | -0.090 |
| rplp2l | 0.128 | uncx | -0.090 |
| rps21 | 0.128 | nsg2 | -0.089 |
| pdia4 | 0.124 | akt1s1 | -0.089 |
| rpl36a | 0.121 | txnipa | -0.088 |
| tmem258 | 0.120 | hpcal4 | -0.088 |
| ppl | 0.118 | cluap1 | -0.087 |
| pdia3 | 0.117 | iqcg | -0.086 |
| agr1 | 0.117 | ip6k2a | -0.086 |
| ndufa2 | 0.116 | gng13b | -0.085 |
| mt-atp8 | 0.116 | elavl3 | -0.085 |
| NDUFB1 | 0.116 | insm1a | -0.085 |
| atp5f1e | 0.115 | gnal | -0.085 |
| capn9 | 0.115 | b3gat1b | -0.084 |
| pycard | 0.114 | si:dkey-276j7.1 | -0.084 |
| zgc:114188 | 0.114 | spag8 | -0.084 |
| rpl35a | 0.114 | pim3 | -0.083 |
| zgc:92380 | 0.114 | tmeff1b | -0.083 |
| rps17 | 0.112 | tcea2 | -0.083 |
| rpl36 | 0.112 | si:dkey-16l2.17 | -0.082 |
| p4hb | 0.112 | rab40b | -0.081 |
| tars | 0.112 | atp6v0cb | -0.081 |
| pmm2 | 0.111 | pacrg | -0.081 |