ORAI calcium release-activated calcium modulator 1b
ZFIN 
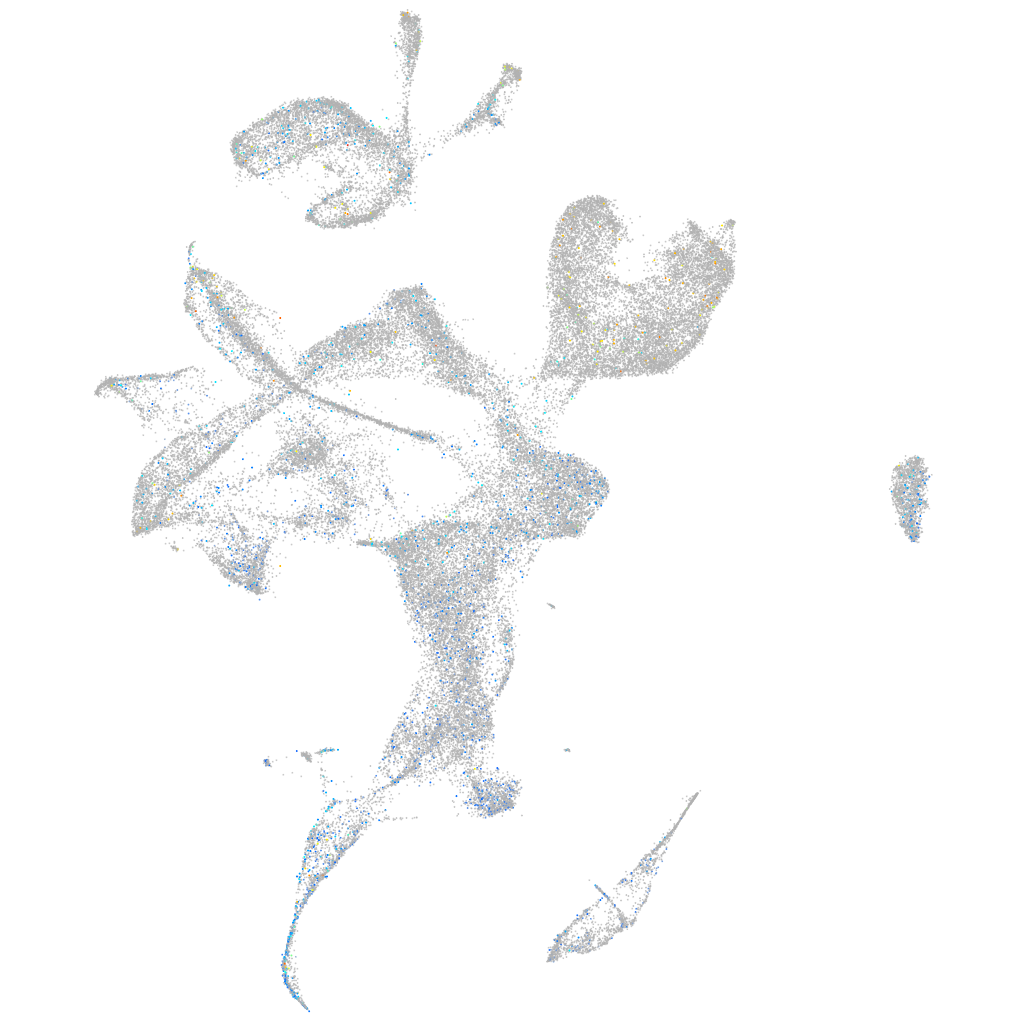
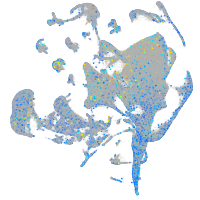
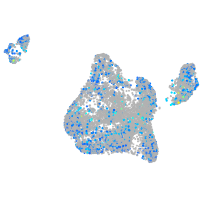
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| pmelb | 0.114 | ptmab | -0.055 |
| tmem98 | 0.107 | si:ch73-1a9.3 | -0.043 |
| tm6sf2 | 0.106 | hmgn6 | -0.038 |
| dct | 0.106 | ptmaa | -0.030 |
| tyrp1b | 0.104 | crx | -0.030 |
| pmela | 0.103 | syt5b | -0.029 |
| msnb | 0.102 | h2afva | -0.028 |
| cracr2ab | 0.102 | epb41a | -0.028 |
| oca2 | 0.102 | rs1a | -0.027 |
| tspan10 | 0.101 | neurod4 | -0.026 |
| rgrb | 0.101 | gnb3a | -0.025 |
| si:ch211-125m10.6 | 0.101 | samsn1a | -0.024 |
| tspan36 | 0.101 | nrn1lb | -0.023 |
| pttg1ipb | 0.100 | mpp6b | -0.023 |
| gpr143 | 0.099 | pcp4l1 | -0.023 |
| tyrp1a | 0.097 | anp32e | -0.022 |
| fabp11b | 0.096 | vsx1 | -0.022 |
| slc45a2 | 0.096 | snap25b | -0.021 |
| sytl2a | 0.096 | gng13b | -0.021 |
| rab27a | 0.095 | h3f3a | -0.021 |
| bace2 | 0.095 | dscaml1 | -0.020 |
| slc7a2 | 0.092 | isl1 | -0.020 |
| sypl1 | 0.092 | hmgn2 | -0.019 |
| fgfbp1a | 0.092 | gpm6aa | -0.019 |
| lrata | 0.092 | hunk | -0.019 |
| stra6 | 0.091 | cadm3 | -0.019 |
| rlbp1b | 0.091 | golga7ba | -0.019 |
| slc24a5 | 0.089 | sypb | -0.019 |
| lpl | 0.089 | nova2 | -0.019 |
| rgra | 0.089 | scrt2 | -0.019 |
| col4a5 | 0.089 | vamp1 | -0.019 |
| rpe65a | 0.089 | cplx4a | -0.018 |
| lratb.2 | 0.088 | calb2b | -0.018 |
| rdh5 | 0.088 | eml1 | -0.018 |
| col18a1a | 0.088 | cabp2a | -0.018 |