opsin 5
ZFIN 
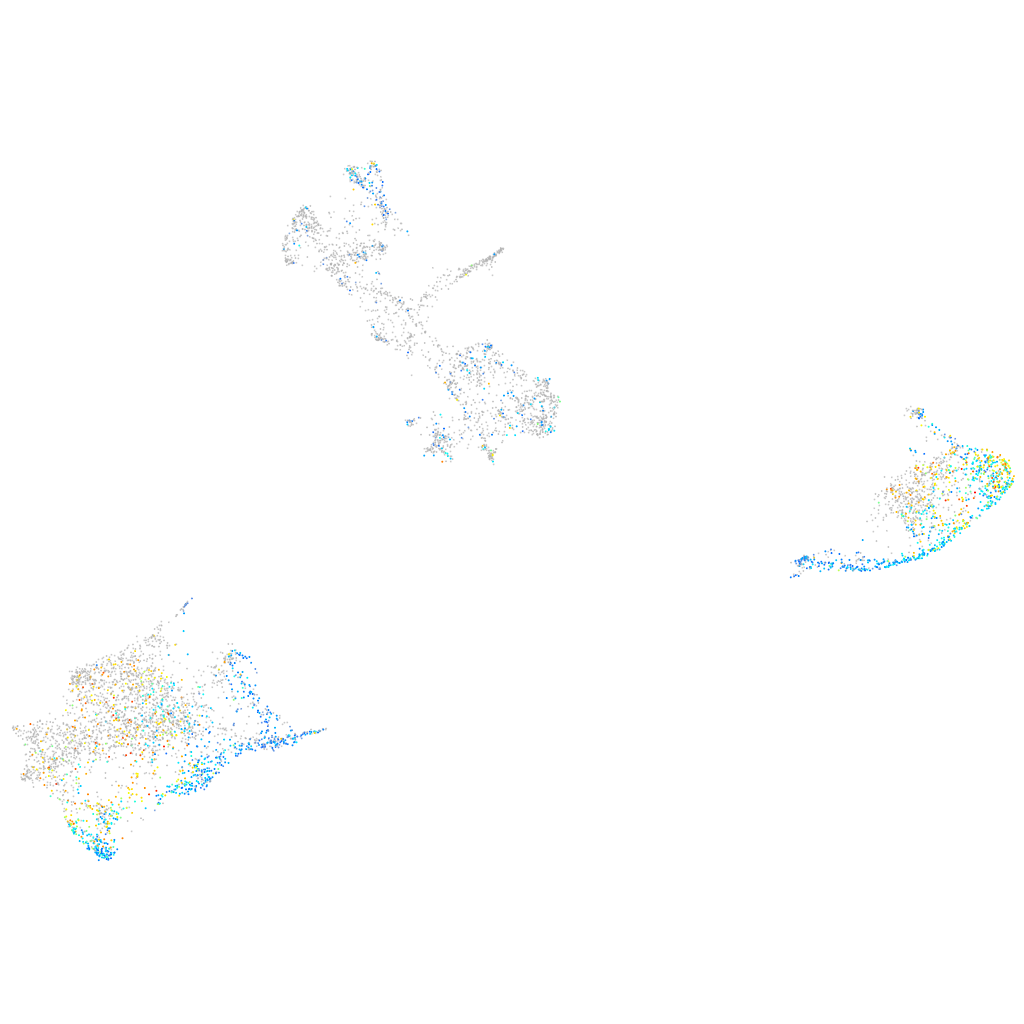
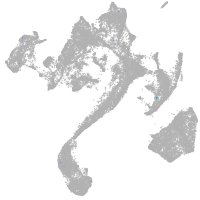
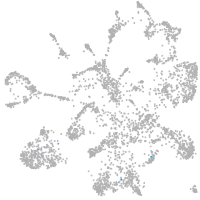
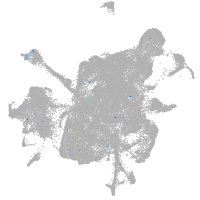
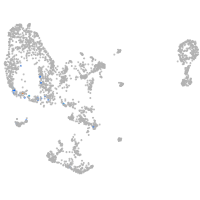
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| kita | 0.372 | defbl1 | -0.213 |
| SPAG9 | 0.356 | apoda.1 | -0.209 |
| mlpha | 0.345 | mdkb | -0.194 |
| kcnj13 | 0.334 | si:ch211-222l21.1 | -0.188 |
| oca2 | 0.330 | si:ch211-256m1.8 | -0.184 |
| tfap2e | 0.316 | gpnmb | -0.180 |
| slc39a10 | 0.313 | si:dkey-197i20.6 | -0.180 |
| bace2 | 0.311 | hmgn2 | -0.176 |
| slc24a5 | 0.311 | lypc | -0.175 |
| pmela | 0.309 | alx4a | -0.168 |
| tyr | 0.308 | ptmab | -0.167 |
| cdh1 | 0.302 | ptmaa | -0.163 |
| tyrp1b | 0.301 | si:ch73-89b15.3 | -0.160 |
| inka1b | 0.298 | unm-sa821 | -0.159 |
| tspan36 | 0.298 | s100v2 | -0.156 |
| tyrp1a | 0.298 | alx1 | -0.151 |
| rasd1 | 0.295 | h2afvb | -0.150 |
| slc22a2 | 0.285 | hmgb1b | -0.149 |
| pcdh10a | 0.280 | si:ch73-1a9.3 | -0.149 |
| si:ch211-241j12.3 | 0.280 | cdh11 | -0.148 |
| dct | 0.279 | akap12a | -0.148 |
| sik1 | 0.279 | h3f3d | -0.147 |
| slc24a4a | 0.279 | guk1a | -0.147 |
| arhgef33 | 0.278 | tuba1b | -0.146 |
| slc3a2a | 0.276 | ponzr1 | -0.145 |
| tspan10 | 0.275 | h3f3a | -0.144 |
| si:ch73-389b16.1 | 0.274 | psat1 | -0.144 |
| lamp1a | 0.274 | slc25a36a | -0.141 |
| zgc:91968 | 0.274 | alx4b | -0.141 |
| myo5aa | 0.271 | hmgb2a | -0.140 |
| mitfa | 0.270 | paics | -0.138 |
| mtbl | 0.267 | hmgb2b | -0.138 |
| si:zfos-943e10.1 | 0.266 | hnrnpaba | -0.138 |
| prkar1b | 0.265 | gapdhs | -0.133 |
| aadac | 0.263 | nova2 | -0.133 |