olfactomedin 2b
ZFIN 
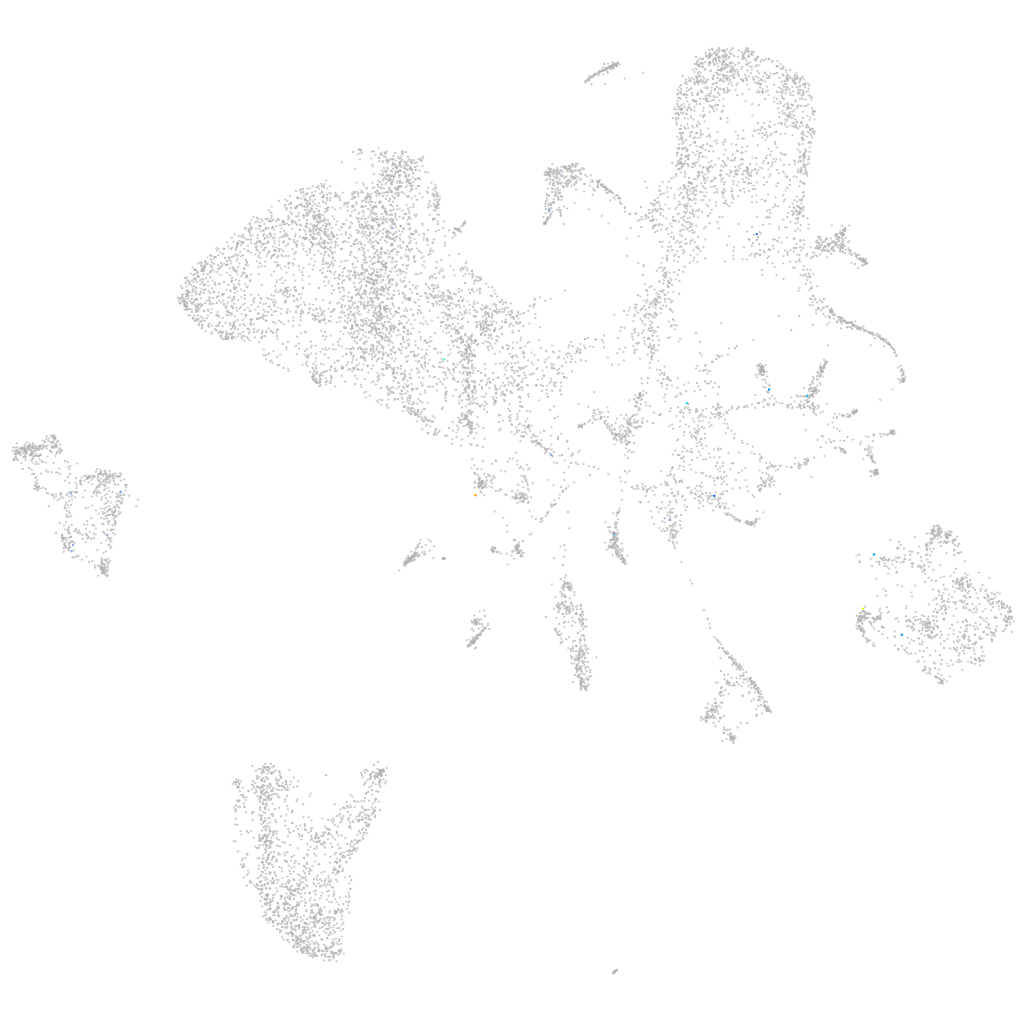
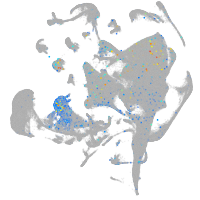
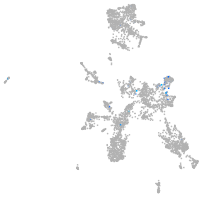
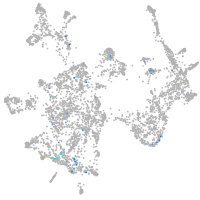
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| wu:fj39g12 | 0.541 | rpl14 | -0.044 |
| si:dkeyp-74b6.2 | 0.428 | uba52 | -0.041 |
| adipoqa | 0.347 | mt-nd4 | -0.039 |
| plppr4b | 0.333 | rps27a | -0.034 |
| znf1043 | 0.319 | nupr1b | -0.034 |
| si:ch211-218d20.15 | 0.316 | gapdh | -0.033 |
| impa2 | 0.298 | aldh7a1 | -0.032 |
| sez6l2 | 0.281 | eef2b | -0.032 |
| ubap1la | 0.267 | rpl4 | -0.032 |
| gabbr2 | 0.265 | cox4i1 | -0.031 |
| neurod2 | 0.264 | rpl36a | -0.031 |
| CR550302.1 | 0.250 | aldob | -0.031 |
| fut9a | 0.240 | gstt1a | -0.031 |
| si:ch73-215f7.1 | 0.222 | glud1b | -0.030 |
| scn1lab | 0.216 | minos1 | -0.029 |
| mag | 0.215 | sec61a1 | -0.029 |
| hpcal4 | 0.210 | rpl26 | -0.028 |
| barhl1b | 0.210 | cx32.3 | -0.028 |
| LOC101882509 | 0.201 | gatm | -0.028 |
| brinp2 | 0.197 | suclg1 | -0.028 |
| dpp6b | 0.195 | atp5mf | -0.028 |
| lingo1a | 0.191 | rps26l | -0.028 |
| CR352342.1 | 0.188 | rplp2l | -0.027 |
| pcnx2 | 0.183 | ndufa4l | -0.027 |
| gpm6ba | 0.176 | p4hb | -0.027 |
| LOC101886256 | 0.170 | cox6b1 | -0.027 |
| jakmip2 | 0.170 | atp5mc3a | -0.027 |
| BX470183.1 | 0.170 | prdx6 | -0.027 |
| kcnj19b | 0.168 | rplp0 | -0.026 |
| tcf4 | 0.166 | cox5ab | -0.026 |
| yjefn3 | 0.165 | ndufa1 | -0.026 |
| zic1 | 0.162 | adka | -0.026 |
| ryr1b | 0.160 | ndufs5 | -0.026 |
| galnt18a | 0.159 | hacd2 | -0.026 |
| LOC100330186 | 0.158 | rps27.2 | -0.026 |