ornithine decarboxylase 1
ZFIN 
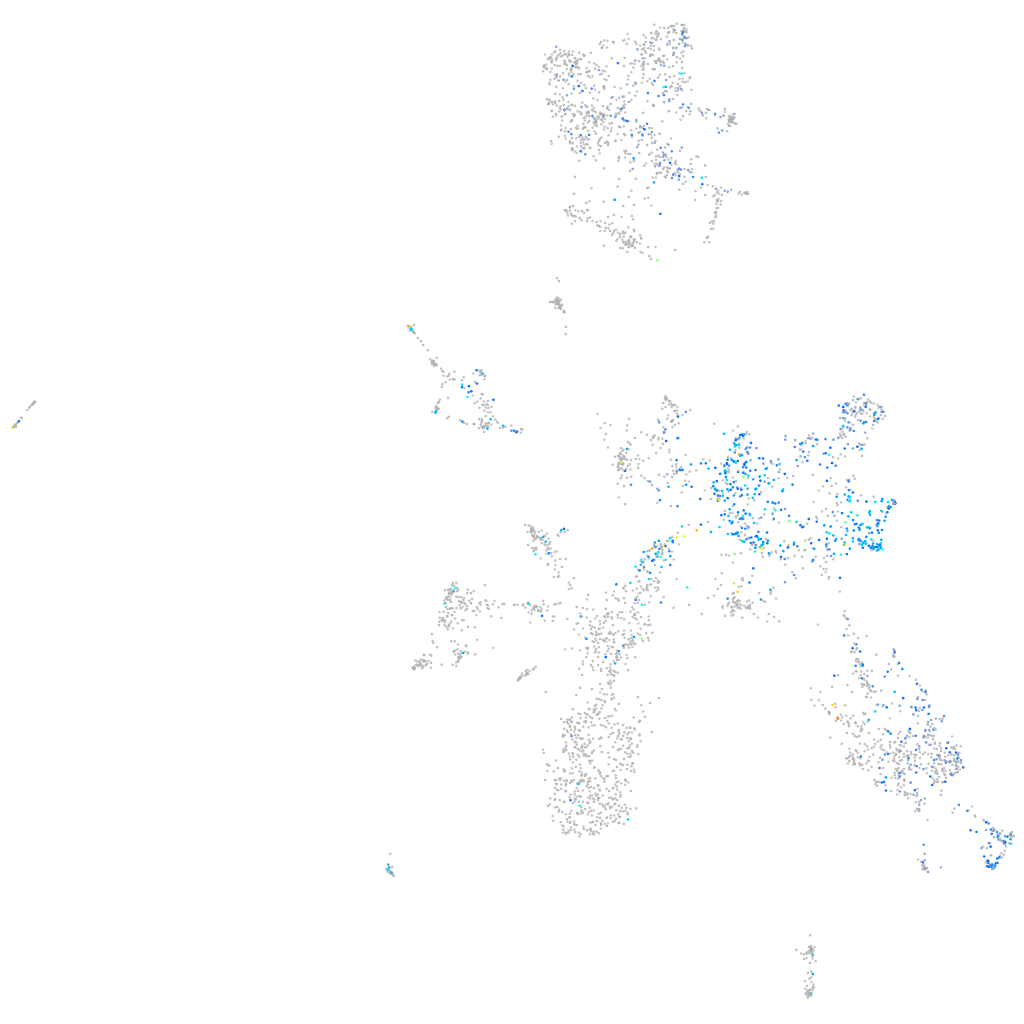
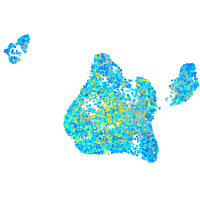
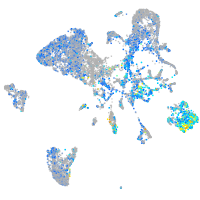
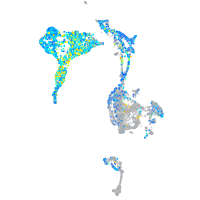
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| her15.1 | 0.423 | gstp1 | -0.335 |
| dlx3b | 0.405 | anxa5b | -0.327 |
| dlx4b | 0.385 | txn | -0.296 |
| hmga1a | 0.384 | icn | -0.265 |
| dut | 0.373 | selenow2b | -0.258 |
| rrm1 | 0.372 | bik | -0.257 |
| fezf1 | 0.371 | cldnh | -0.252 |
| XLOC-003692 | 0.371 | wu:fb18f06 | -0.250 |
| her4.2 | 0.361 | fxyd1 | -0.249 |
| stmn1a | 0.355 | tmem59 | -0.246 |
| dla | 0.353 | gapdhs | -0.238 |
| her2 | 0.350 | COX3 | -0.235 |
| ranbp1 | 0.347 | calm1a | -0.223 |
| ccna2 | 0.346 | calb2a | -0.222 |
| cks1b | 0.346 | calm1b | -0.221 |
| zgc:110216 | 0.344 | nfe2l2a | -0.220 |
| hnrnpabb | 0.344 | gnb1a | -0.218 |
| cdca5 | 0.343 | selenow1 | -0.217 |
| nasp | 0.343 | vamp2 | -0.215 |
| cx43.4 | 0.339 | pvalb5 | -0.215 |
| chaf1a | 0.337 | ponzr6 | -0.214 |
| si:ch73-281n10.2 | 0.336 | gsto2 | -0.210 |
| her12 | 0.335 | gbgt1l4 | -0.208 |
| rrm2 | 0.335 | rnasekb | -0.207 |
| hmgb2b | 0.333 | TSTD1 | -0.201 |
| si:ch211-222l21.1 | 0.333 | tnks1bp1 | -0.200 |
| ran | 0.332 | si:ch211-153b23.5 | -0.200 |
| cdca7b | 0.329 | uchl1 | -0.196 |
| ube2c | 0.328 | htatip2 | -0.194 |
| mad2l1 | 0.326 | ap1s3b | -0.194 |
| mki67 | 0.326 | si:ch1073-443f11.2 | -0.193 |
| mcm7 | 0.325 | chga | -0.193 |
| CU467822.1 | 0.324 | gabarapa | -0.192 |
| setb | 0.323 | mt-co2 | -0.192 |
| her4.1 | 0.323 | flrt1b | -0.192 |