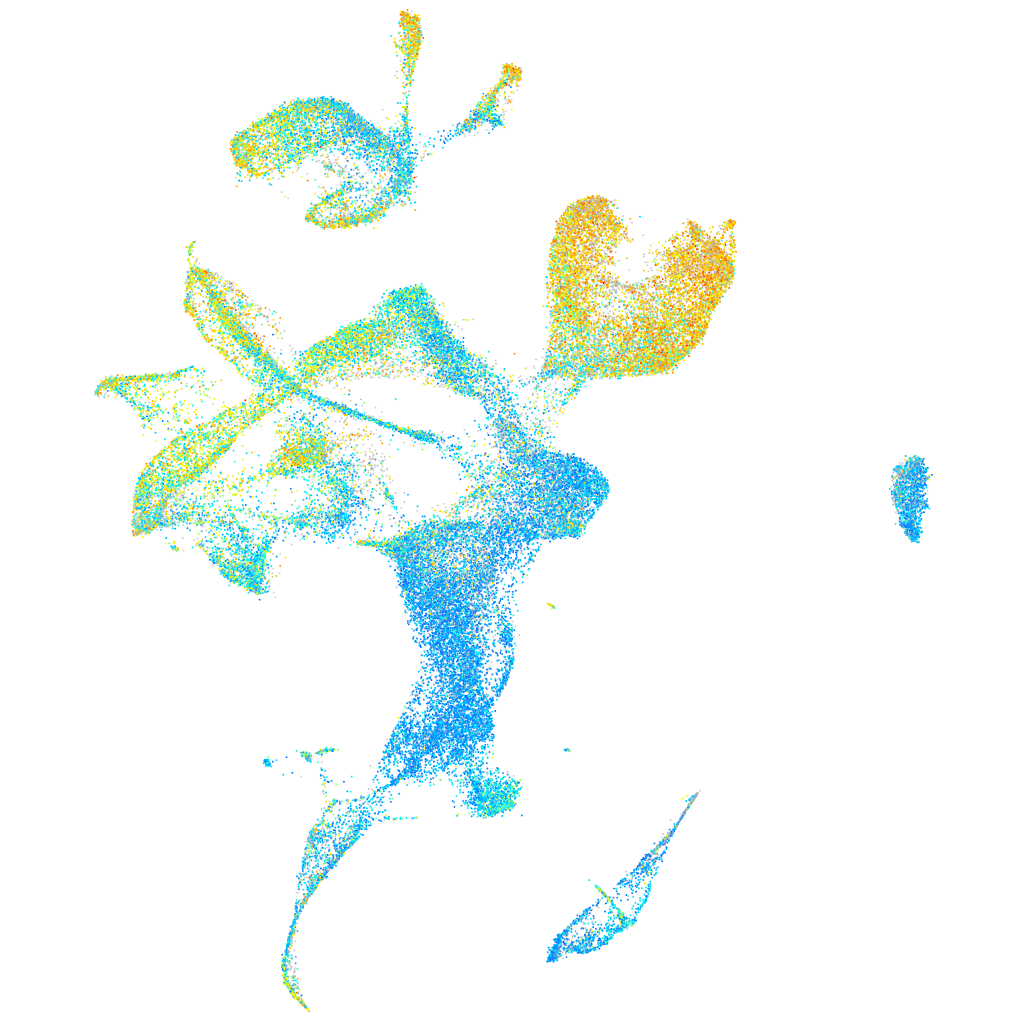ornithine decarboxylase antizyme 1a
ZFIN 
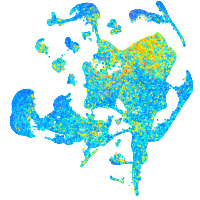
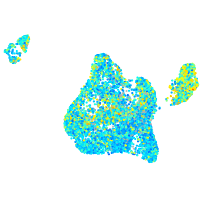
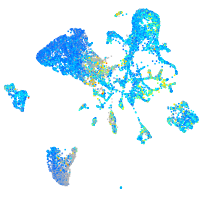
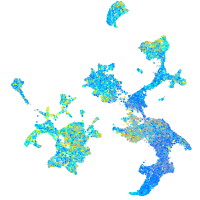
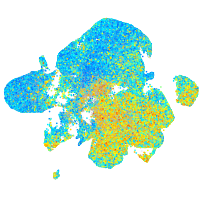
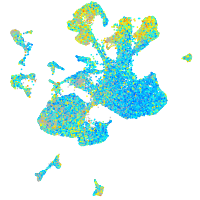
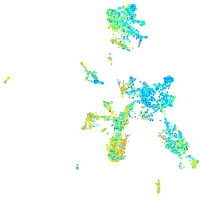
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| atp6v0cb | 0.316 | rpl9 | -0.232 |
| atp6v1e1b | 0.307 | pcna | -0.228 |
| snap25b | 0.305 | si:ch211-222l21.1 | -0.226 |
| ckbb | 0.301 | stmn1a | -0.218 |
| atp6v1g1 | 0.297 | rrm1 | -0.216 |
| rnasekb | 0.295 | rplp2l | -0.215 |
| gapdhs | 0.282 | rplp1 | -0.211 |
| sypb | 0.270 | si:dkey-151g10.6 | -0.210 |
| atpv0e2 | 0.264 | lbr | -0.208 |
| calm1a | 0.256 | ccnd1 | -0.204 |
| oaz1b | 0.251 | mki67 | -0.203 |
| atp6ap2 | 0.246 | banf1 | -0.201 |
| tpi1b | 0.241 | rpl38 | -0.199 |
| syt5b | 0.241 | mdka | -0.199 |
| rs1a | 0.236 | rps28 | -0.198 |
| si:ch1073-429i10.3.1 | 0.232 | mcm7 | -0.197 |
| sncb | 0.222 | chaf1a | -0.196 |
| eif1b | 0.222 | lig1 | -0.196 |
| ndrg4 | 0.222 | rrm2 | -0.195 |
| h3f3c | 0.219 | tuba8l4 | -0.195 |
| atp6v1h | 0.218 | dut | -0.193 |
| vamp2 | 0.218 | zgc:165555.3 | -0.193 |
| gng3 | 0.217 | rps12 | -0.193 |
| gng13b | 0.213 | rpl29 | -0.193 |
| ppdpfb | 0.213 | CABZ01005379.1 | -0.191 |
| cnrip1b | 0.213 | hmgb2b | -0.191 |
| olfm1b | 0.212 | ccna2 | -0.190 |
| gnao1b | 0.210 | rpl4 | -0.189 |
| lin7a | 0.207 | msna | -0.188 |
| necap1 | 0.205 | hes2.2 | -0.187 |
| calb2b | 0.203 | nasp | -0.187 |
| gnb5b | 0.202 | si:dkey-261m9.17 | -0.187 |
| atp1b2a | 0.202 | tuba8l | -0.186 |
| mpp6b | 0.201 | dek | -0.186 |
| gabarapb | 0.199 | hmga1a | -0.185 |