NUS1 dehydrodolichyl diphosphate synthase subunit
ZFIN 
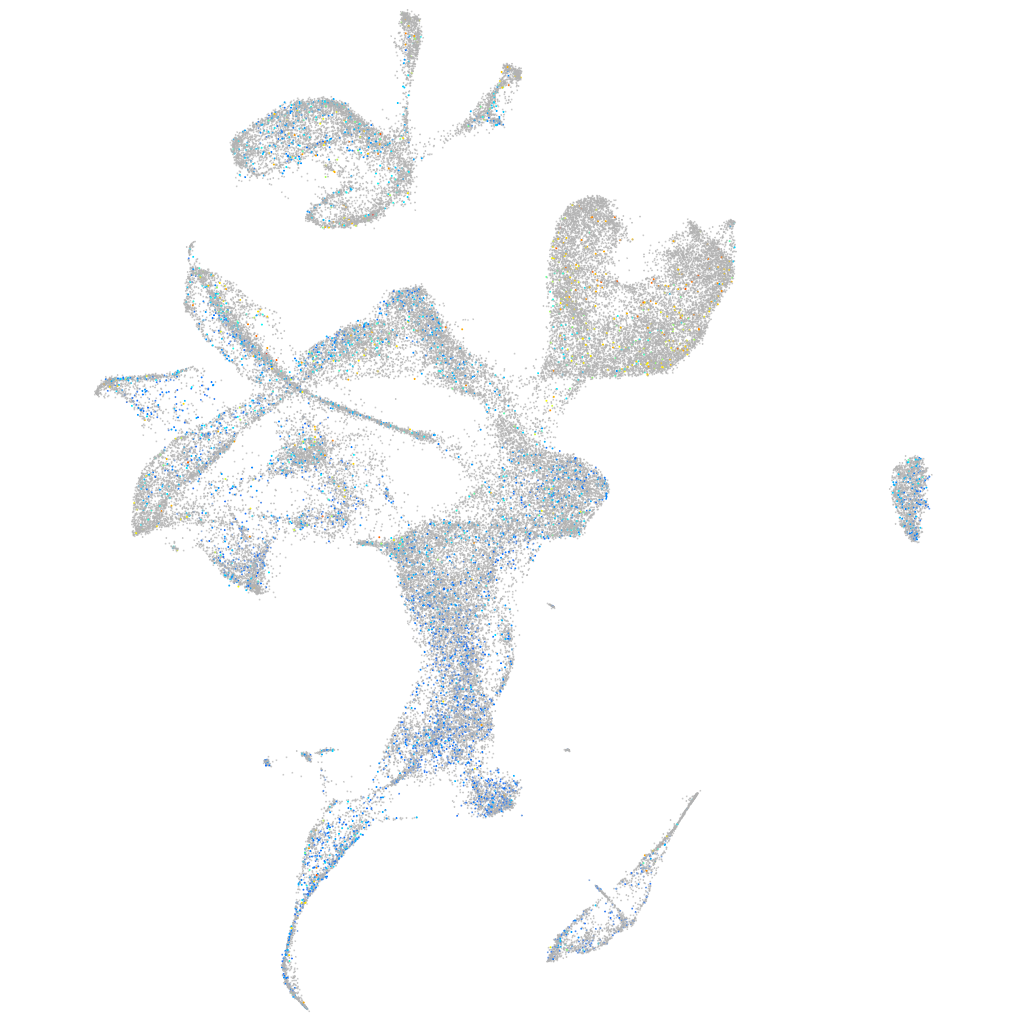
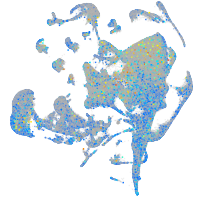
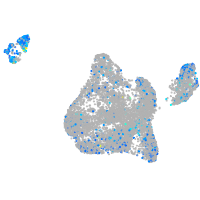
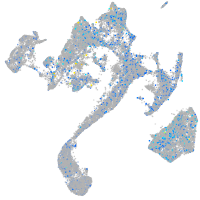
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| rab32a | 0.065 | pvalb2 | -0.042 |
| hspa5 | 0.064 | pvalb1 | -0.042 |
| calr3b | 0.063 | actc1b | -0.041 |
| tyrp1a | 0.060 | mylpfa | -0.032 |
| zgc:110591 | 0.060 | si:ch73-256g18.2 | -0.032 |
| slc45a2 | 0.060 | tnni2a.4 | -0.032 |
| gpr143 | 0.058 | apoa2 | -0.031 |
| tspan36 | 0.058 | cabp2a | -0.030 |
| pmelb | 0.058 | mylz3 | -0.030 |
| slc24a5 | 0.057 | nrn1lb | -0.029 |
| calr | 0.057 | si:dkey-16p21.8 | -0.029 |
| hsp90b1 | 0.057 | apoa1b | -0.027 |
| tyr | 0.057 | cplx4a | -0.026 |
| canx | 0.057 | prss1 | -0.026 |
| rab5if | 0.056 | myl1 | -0.025 |
| tyrp1b | 0.056 | arhgef25b | -0.025 |
| FP085398.1 | 0.056 | tnnt3b | -0.025 |
| ppib | 0.056 | tpma | -0.025 |
| col4a6 | 0.056 | CELA1 (1 of many) | -0.025 |
| rab38 | 0.056 | calb2b | -0.024 |
| dct | 0.055 | vamp1 | -0.024 |
| mitfa | 0.055 | prss59.2 | -0.024 |
| ssr4 | 0.055 | ctrb1 | -0.024 |
| qdpra | 0.054 | rgs16 | -0.023 |
| cracr2ab | 0.054 | ptmab | -0.023 |
| ssr2 | 0.054 | myhz1.1 | -0.023 |
| sytl2b | 0.054 | gapdh | -0.023 |
| pdia3 | 0.054 | si:ch1073-83n3.2 | -0.023 |
| sptlc2a | 0.054 | prss59.1 | -0.022 |
| pmela | 0.054 | dusp2 | -0.022 |
| agtrap | 0.053 | snx8b | -0.022 |
| pnp4a | 0.053 | rgs11 | -0.021 |
| oca2 | 0.053 | ip6k2a | -0.021 |
| tspan10 | 0.052 | krt4 | -0.021 |
| col4a5 | 0.052 | cabp5b | -0.020 |