nucleobindin 2b
ZFIN 
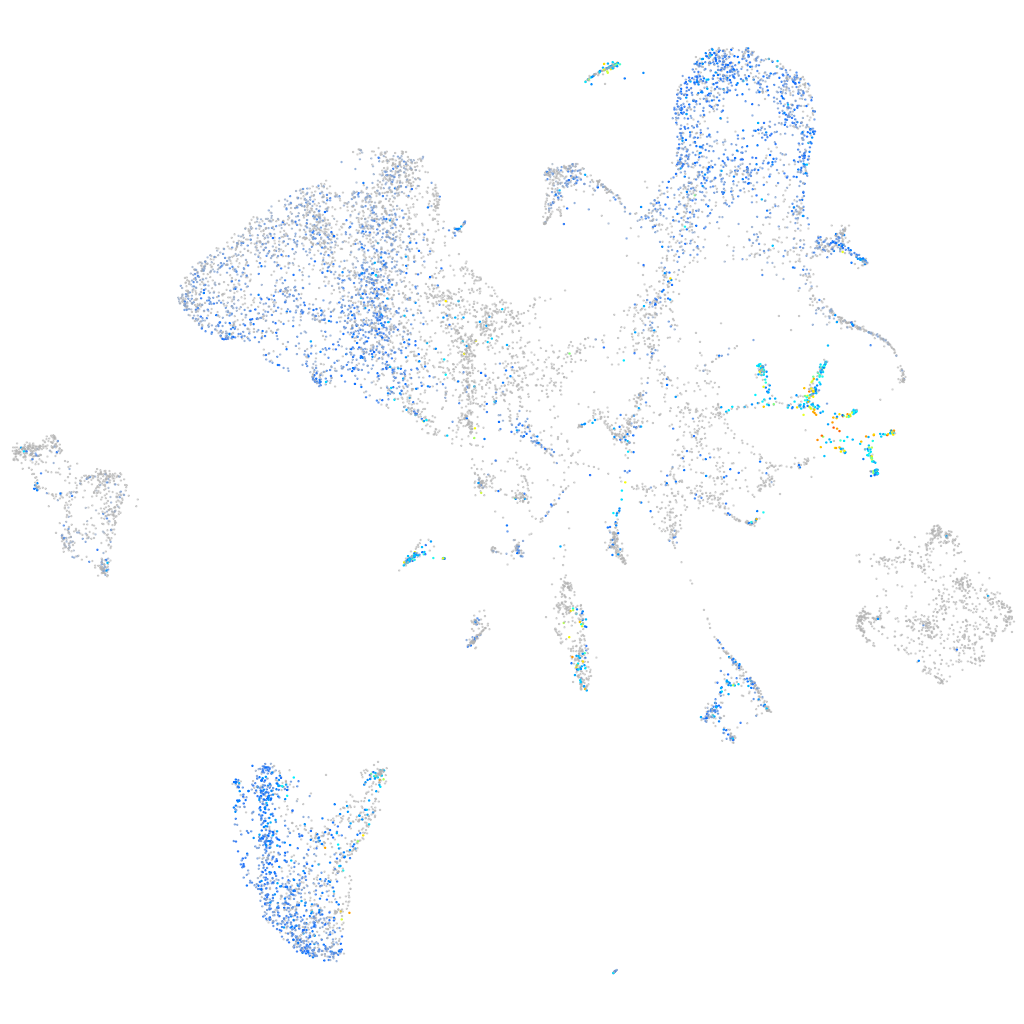
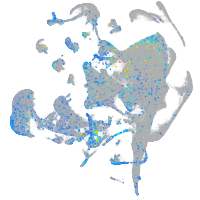
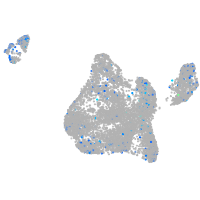
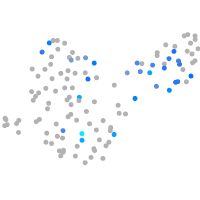
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| scg3 | 0.468 | apoc1 | -0.176 |
| neurod1 | 0.422 | cx43.4 | -0.161 |
| scgn | 0.418 | id1 | -0.158 |
| rprmb | 0.384 | hspb1 | -0.151 |
| mir7a-1 | 0.379 | qkia | -0.149 |
| pcsk1nl | 0.378 | stmn1a | -0.143 |
| pax6b | 0.371 | id3 | -0.141 |
| si:dkey-153k10.9 | 0.358 | akap12b | -0.141 |
| kcnj11 | 0.355 | parp1 | -0.136 |
| c2cd4a | 0.355 | mcm6 | -0.134 |
| nkx2.2a | 0.347 | marcksb | -0.134 |
| pcsk2 | 0.342 | cdh6 | -0.133 |
| isl1 | 0.336 | si:ch211-152c2.3 | -0.132 |
| insm1a | 0.334 | lig1 | -0.130 |
| slc1a4 | 0.332 | hnrnpa1b | -0.124 |
| mir375-2 | 0.331 | apoeb | -0.123 |
| syt1a | 0.328 | ppig | -0.121 |
| pcsk1 | 0.325 | hmgb2b | -0.120 |
| ptprna | 0.325 | dkc1 | -0.120 |
| arxa | 0.325 | anp32b | -0.120 |
| tspan7b | 0.321 | hnrnpm | -0.120 |
| fev | 0.320 | cbx1a | -0.119 |
| rasd4 | 0.318 | fabp10a | -0.119 |
| hepacam2 | 0.317 | cdca7a | -0.118 |
| scg2a | 0.317 | nasp | -0.117 |
| gpc1a | 0.314 | cdca7b | -0.117 |
| atp6v0cb | 0.313 | zfp36l1a | -0.117 |
| kcnk9 | 0.310 | slc16a1b | -0.117 |
| cacna1c | 0.306 | smo | -0.117 |
| surf4l | 0.305 | mki67 | -0.117 |
| tango2 | 0.305 | seta | -0.116 |
| vat1 | 0.304 | wu:fb97g03 | -0.116 |
| zgc:101731 | 0.303 | cxcr4a | -0.116 |
| elovl1a | 0.292 | rps12 | -0.116 |
| rims2a | 0.290 | tgif1 | -0.116 |