"nuclear receptor subfamily 6, group A, member 1a"
ZFIN 
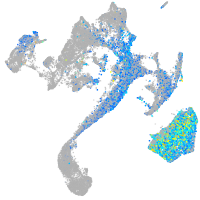
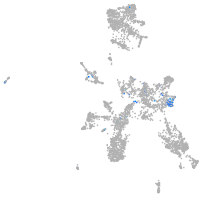
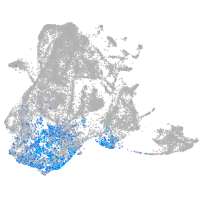
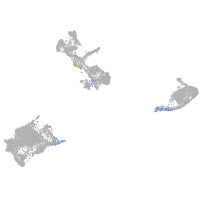
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| hspb1 | 0.621 | rpl37 | -0.294 |
| shisa2a | 0.570 | ptmaa | -0.289 |
| stm | 0.555 | rps10 | -0.283 |
| si:ch211-152c2.3 | 0.551 | h3f3a | -0.275 |
| apoeb | 0.515 | CR383676.1 | -0.263 |
| tdgf1 | 0.504 | zgc:114188 | -0.249 |
| sox19a | 0.500 | ppiab | -0.230 |
| hesx1 | 0.492 | hnrnpa0l | -0.215 |
| cyp26a1 | 0.491 | gpm6aa | -0.211 |
| aldob | 0.470 | marcksl1a | -0.208 |
| zmp:0000000624 | 0.455 | rps17 | -0.186 |
| COX7A2 | 0.443 | COX3 | -0.183 |
| NC-002333.4 | 0.440 | zgc:158463 | -0.181 |
| s100a1 | 0.439 | tuba1c | -0.178 |
| chrd | 0.428 | si:ch1073-429i10.3.1 | -0.169 |
| pprc1 | 0.424 | h3f3c | -0.165 |
| alcamb | 0.412 | fabp3 | -0.161 |
| apoc1 | 0.405 | mt-atp6 | -0.159 |
| LOC108190024 | 0.401 | zgc:153409 | -0.158 |
| gnl3 | 0.399 | pvalb1 | -0.155 |
| nop2 | 0.398 | mt-nd1 | -0.155 |
| bms1 | 0.396 | pvalb2 | -0.154 |
| mybbp1a | 0.391 | mt-co2 | -0.152 |
| npm1a | 0.390 | cadm3 | -0.152 |
| pkdccb | 0.389 | gpm6ab | -0.150 |
| fbl | 0.387 | ckbb | -0.149 |
| asb11 | 0.383 | actc1b | -0.149 |
| foxd5 | 0.378 | celf2 | -0.140 |
| gar1 | 0.376 | ppdpfb | -0.135 |
| dkc1 | 0.373 | hbbe1.3 | -0.134 |
| polr3gla | 0.370 | rtn1a | -0.129 |
| rsl1d1 | 0.368 | sinhcafl | -0.127 |
| nop58 | 0.368 | stmn1b | -0.127 |
| pes | 0.366 | fabp7a | -0.126 |
| dnmt3bb.2 | 0.366 | hbae3 | -0.125 |